Search Count: 1,929
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: CA, 117, NA, MLI, ACY, SIN, FMT |
 |
Crystal Structure Of Keap1 In Complex With A Small Molecule Inhibitor, Kmn003
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: PEPTIDE BINDING PROTEIN/INHIBITOR Ligands: A1L98, FMT |
 |
Organism: Burkholderia cenocepacia h111
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN Ligands: FE, ACY, FMT, NA |
 |
Organism: Streptococcus intermedius
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: FMT, ACY |
 |
Crystal Structure Of Streptomyces Sp. Sulfotransferase Cpz4 Involved In Caprazamycins Synthesis
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, MG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: FMT, GOL |
 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: A1CMH, EDO, FMT |
 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: A1CMG, EDO, FMT |
 |
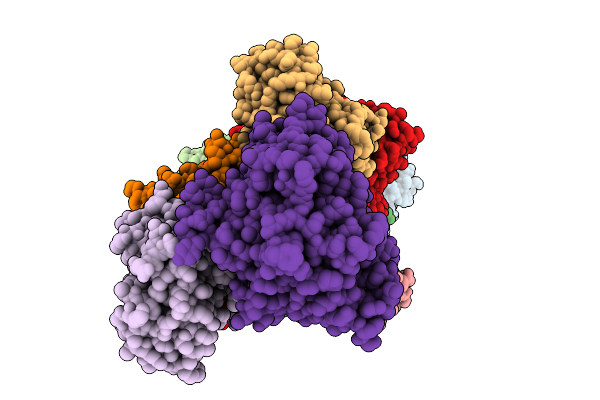
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: TRANSCRIPTION Ligands: A1L9D, FMT |
 |
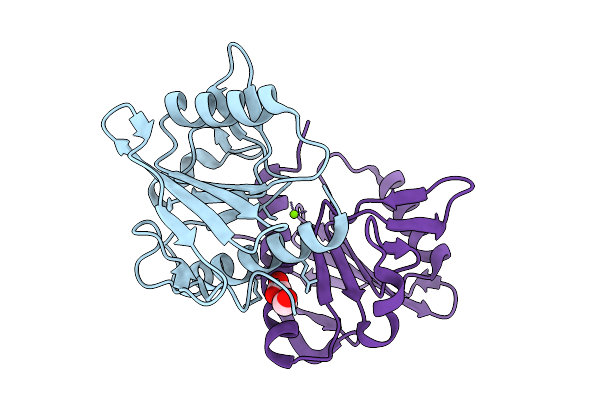
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: ONCOPROTEIN Ligands: GDP, MG, FMT, MLA, ACT |
 |
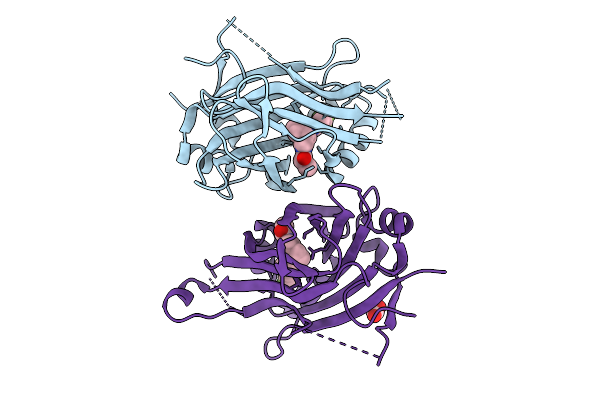
Crystal Structure Of Putative Restriction Endonuclease Domain-Containing Protein From Leptospirillum Ferriphilum Ysk
Organism: Leptospirillum ferriphilum ysk
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE Ligands: FMT, MG, CL |
 |
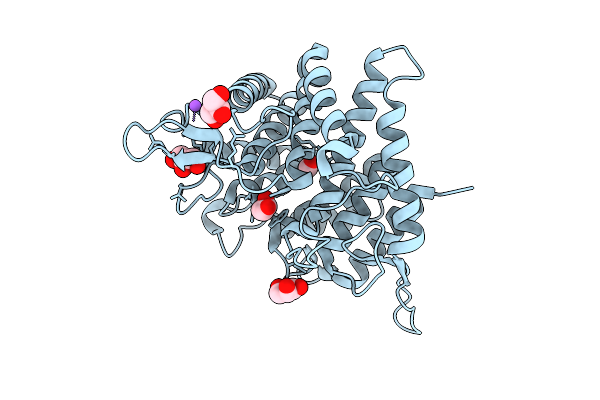
Crystal Structure Of Human Tead2-Yap Binding Domain Covalently Bound To An Allosteric Regulator
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSCRIPTION/INHIBITOR Ligands: EI6, FMT |
 |
Organism: Thermochaetoides thermophila dsm 1495
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: GOL, FMT, NA |
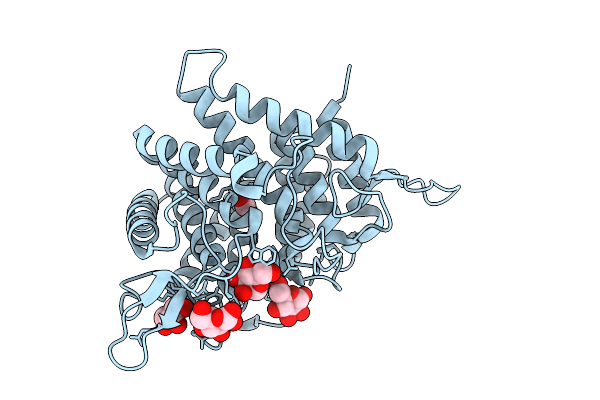 |
Crystal Structure Of Ctgh76 From Chaetomium Thermophilum In Complex With Mannose
Organism: Thermochaetoides thermophila dsm 1495
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: GOL, FMT, MAN |
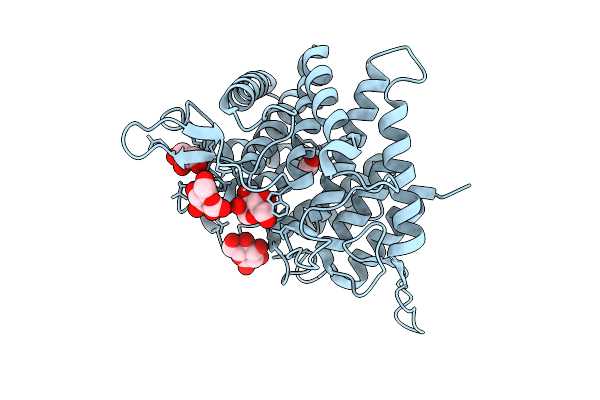 |
Crystal Structure Of Ctgh76 From Chaetomium Thermophilum In Complex With Mannose Products Form
Organism: Thermochaetoides thermophila dsm 1495
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: MAN, GOL, FMT |
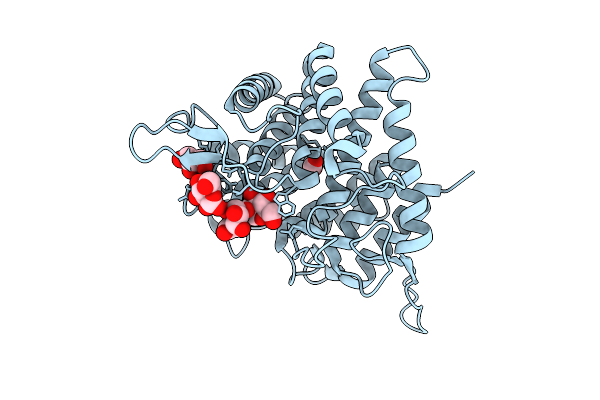 |
Organism: Thermochaetoides thermophila dsm 1495
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: MAN, GOL, FMT |
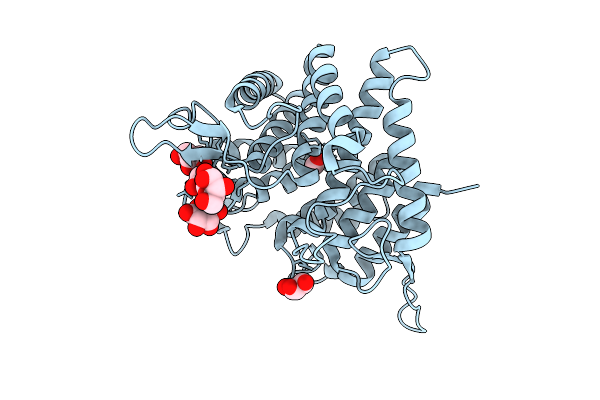 |
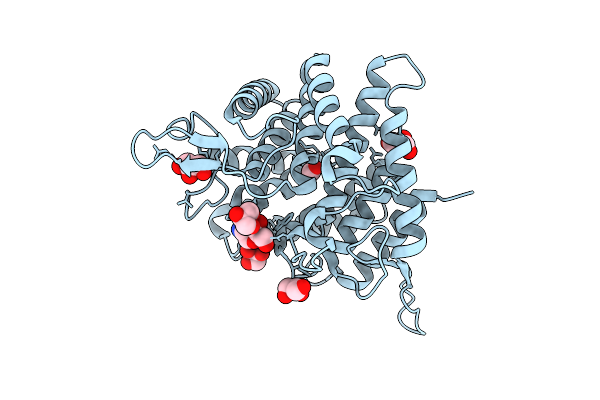
Crystal Structure Of Ctgh76 From Chaetomium Thermophilum In Complex With Alpha-1,3-1,6-Mannotriose
Organism: Thermochaetoides thermophila dsm 1495
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: GOL, FMT |
 |
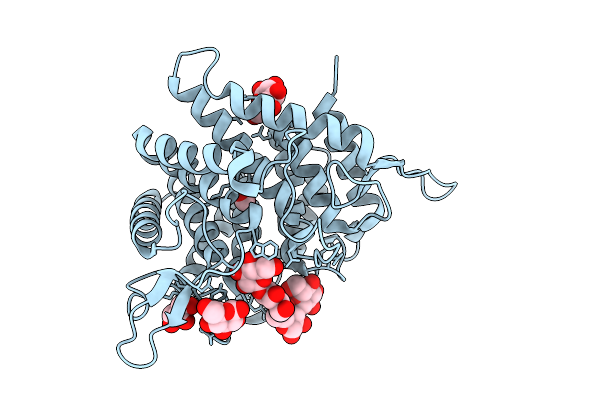
Crystal Structure Of Ctgh76 From Chaetomium Thermophilum In Complex With Glucosamine
Organism: Thermochaetoides thermophila dsm 1495
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: PA1, GOL, FMT |
 |
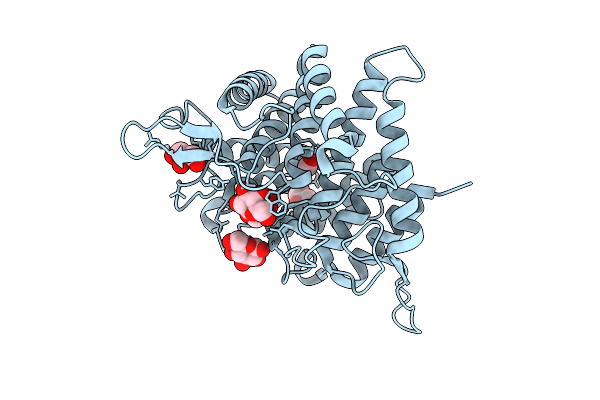
Crystal Structure Of Ctgh76 From Chaetomium Thermophilum In Complex With Laminaribiose
Organism: Thermochaetoides thermophila dsm 1495
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: BGC, GOL, FMT |
 |
Crystal Structure Of Ctgh76 From Chaetomium Thermophilum In Complex With Isomaltose
Organism: Thermochaetoides thermophila dsm 1495
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: BGC, GOL, FMT |

