Search Count: 1,936
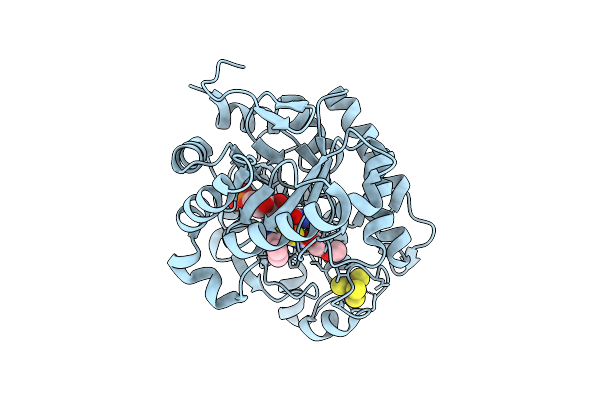 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 1
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, A1ELE, FMN |
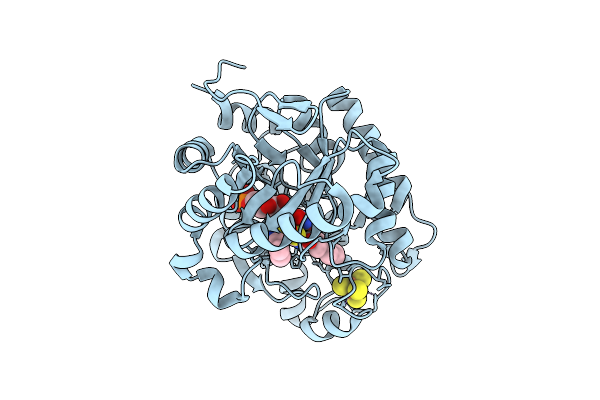 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 21
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1ELF |
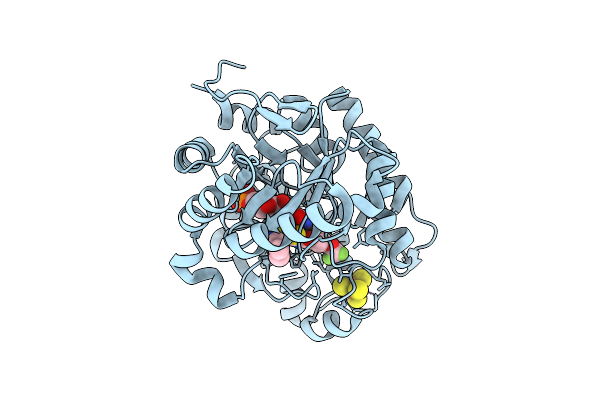 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 11
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1ELG |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 35
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1ELH |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 47
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1EMD |
 |
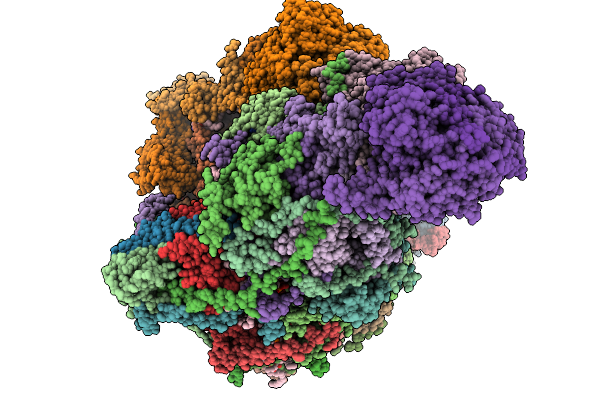
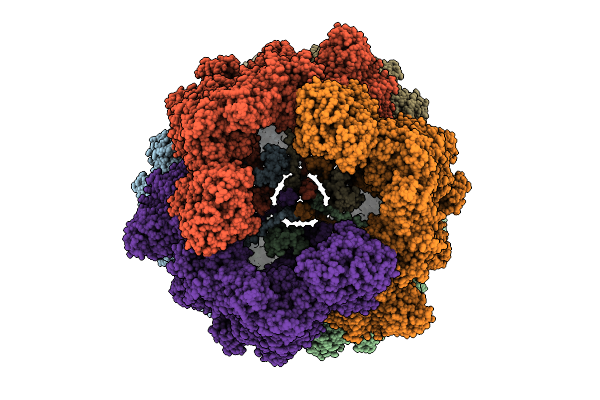
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: CDL, PEE, NDP, ZMP, ADP, 3PE, PC1, PLX, XEW, SF4, FES, MG, ZN, FMN |
 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN |
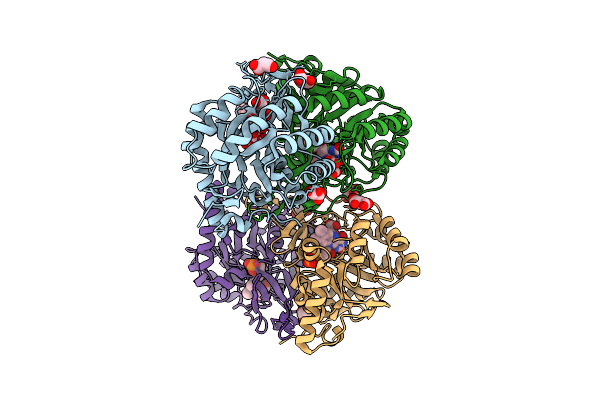 |
Crystal Structure Of The C131A Mutant Of Trypanosoma Brucei Dhodh In Fmn-Oxidized, Ligand-Free Form
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN Ligands: FMN, MLI |
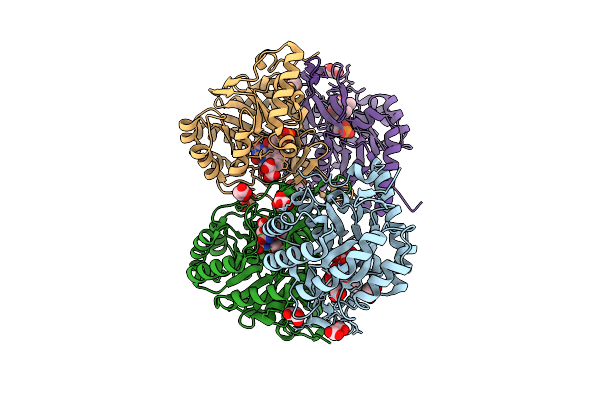 |
Crystal Structure Of The C131A Mutant Of Trypanosoma Brucei Dhodh In Fmn-Oxidized, Dihydroorotate-Bound Form
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN Ligands: FMN, DOR, MLI |
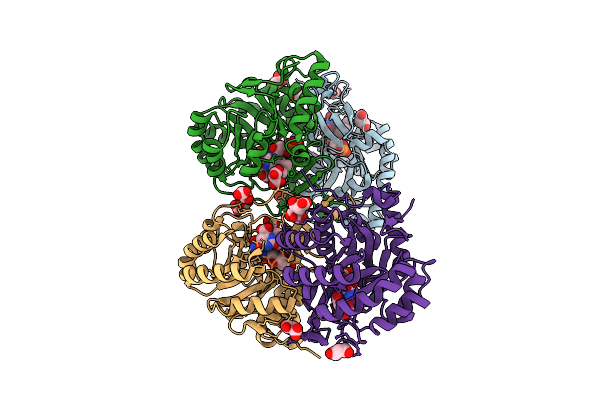 |
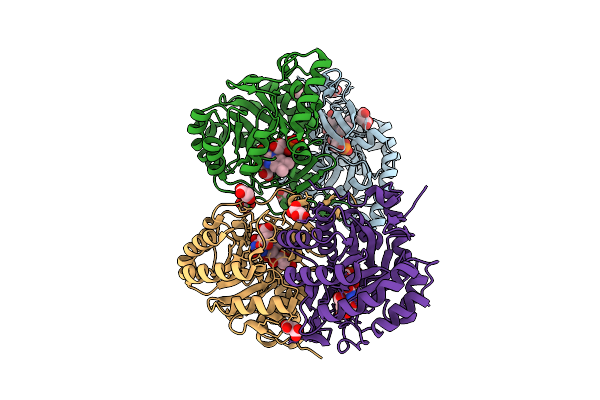
Crystal Structure Of The C131A Mutant Of Trypanosoma Brucei Dhodh In Fmn-Oxidized, Orotate-Bound Form
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN Ligands: FMN, ORO, MLI |
 |
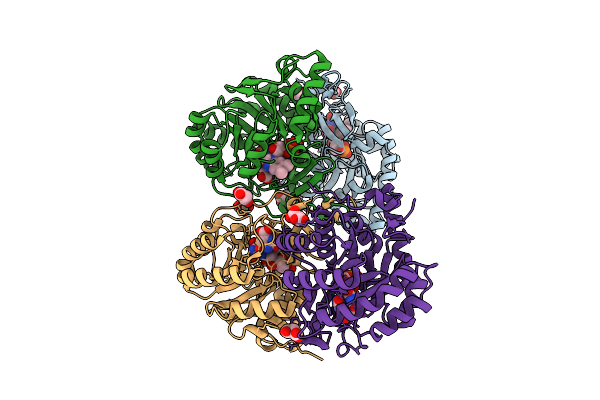
Crystal Structure Of The C131A Mutant Of Trypanosoma Brucei Dhodh In Fmn-Oxidized, Fumarate-Bound Form
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN Ligands: FMN, FUM, MLI |
 |
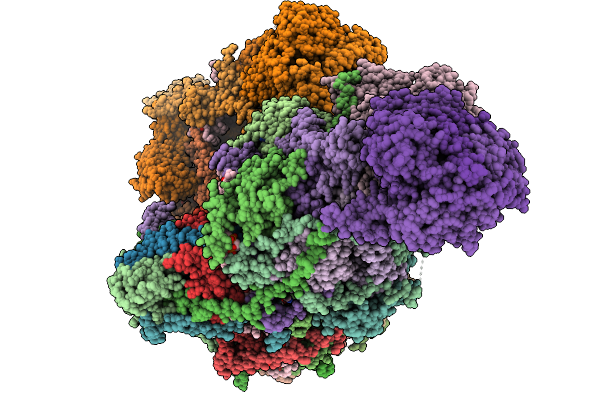
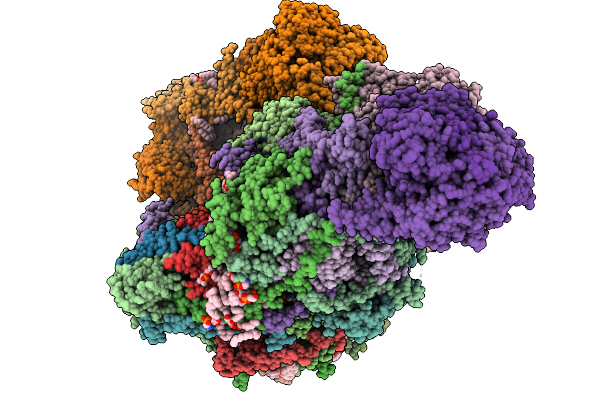
Crystal Structure Of Wild-Type Trypanosoma Brucei Dhodh In Fmn-Oxidized, Orotate-Bound Form
Organism: Trypanosoma brucei brucei treu927
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: FLAVOPROTEIN Ligands: FMN, ORO, MLI |
 |
 |
 |
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10, Alternative Orientation (Sc-Metc1-Ii)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, PX2, U10, SF4, FES, MG, MF8, ZN, FMN |
 |
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10 (Sc-Metc1-I)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, U10, SF4, FES, MG, MF8, ZN, FMN |
 |
Complex I From Respirasome Closed State 1 Bound By Metformin (Sc-Metc1-Iii)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, SF4, FES, MG, MF8, ZN, FMN |
 |