Search Count: 37,671
 |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: CDL, PEE, NDP, ZMP, ADP, 3PE, PC1, PLX, XEW, SF4, FES, MG, ZN, FMN |
 |
Crystal Structure Of S. Thermophilus Class Iii Ribonucleotide Reductase Bound To Datp
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: MG, DTP, ZN, SO4 |
 |
Organism: Streptococcus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: TTP, DTP, MG, ZN |
 |
Organism: Streptococcus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: ATP, MG, TTP, ZN |
 |
Crystal Structure Of C36S Mutant Glutathione Peroxidase Of Staphylococcus Aureus.
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE |
 |
Organism: Komagataella pastoris
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP |
 |
Organism: Komagataella pastoris
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Komagataella pastoris
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP |
 |
Cryo-Em Structure Of Soluble Methane Monooxygenase Hydroxylase From Methylosinus Sporium 5
Organism: Methylosinus sporium
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: FLAVOPROTEIN |
 |
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: FLAVOPROTEIN |
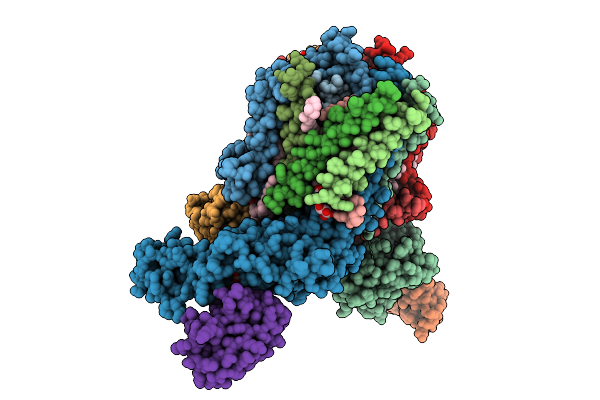 |
Cryo-Em Structure Of Spinacia Oleracea Cytochrome B6F Complex With Bound Plastocyanin
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: HEM, HEC, UMQ, PL9, CLA, FES, SQD, BCR, CU |
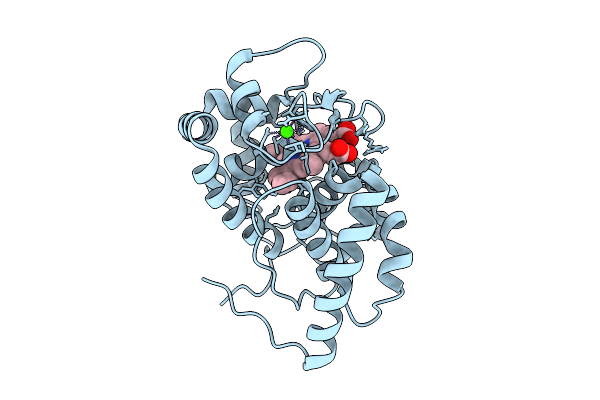 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: HEM, CA |
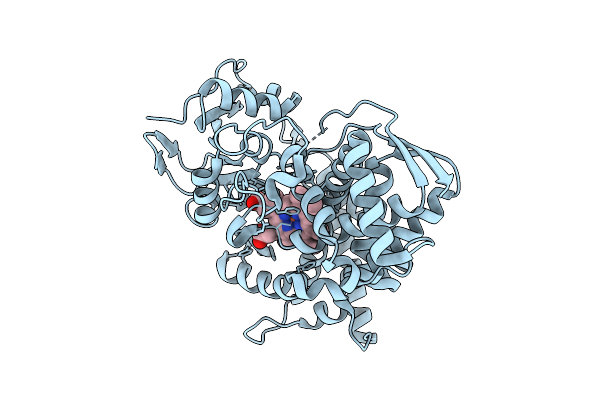 |
Organism: Novosphingobium sp. 28-62-57
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: HEM |
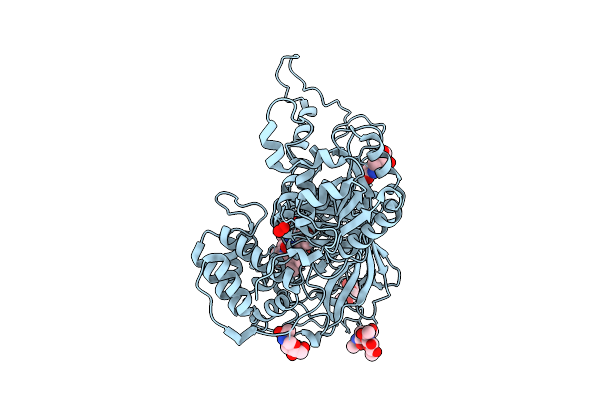 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: HEM, NAG, PEO, CL |
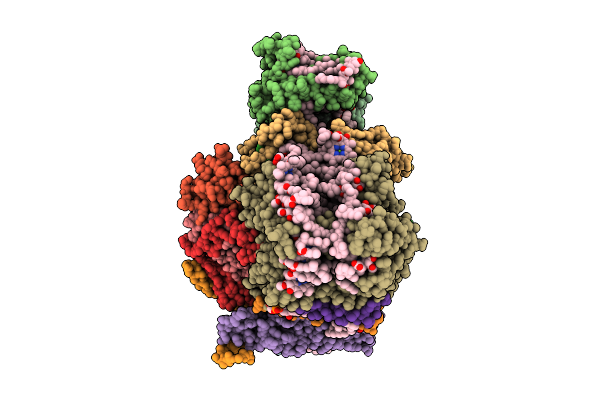 |
Structure Of The Wild-Type Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
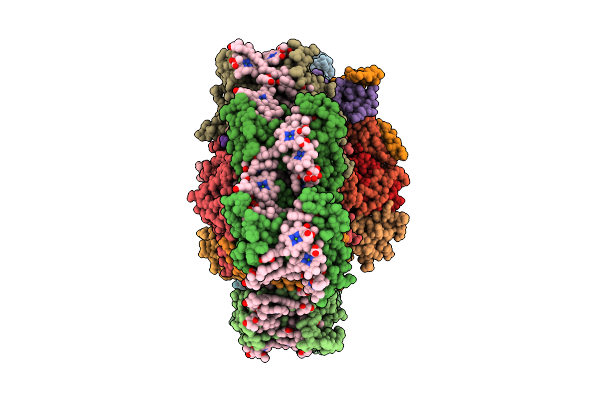 |
Structure Of The Canthaxanthin Mutant Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, 45D, LHG, DGD, LMG, PQN, BCR, SF4 |
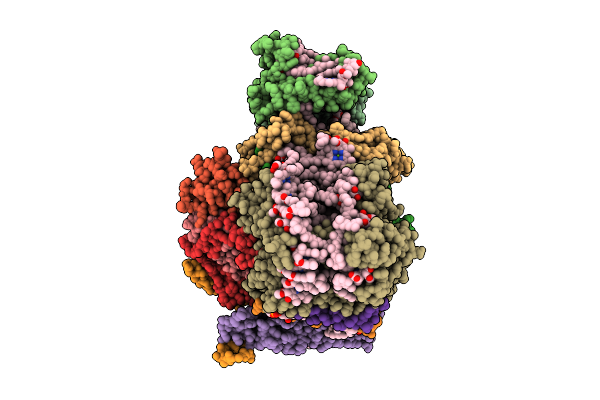 |
Structure Of The Canthaxanthin Mutant Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
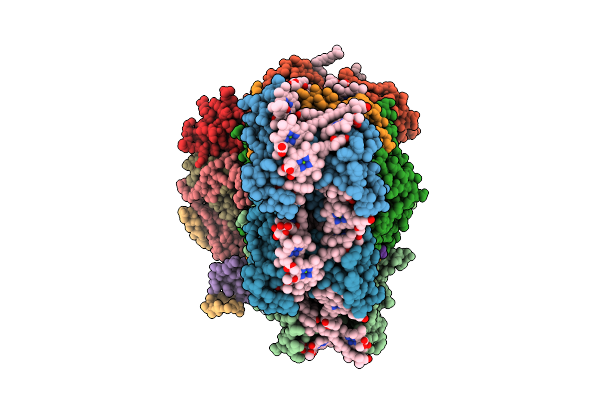 |
Structure Of The Canthaxanthin Mutant Psi-4Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: A1L1G, A1L1F, XAT, 45D, LHG, CLA, DGD, SQD, PQN, BCR, SF4, LMG |

