Search Count: 200
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: FAD, NAP |
 |
Mycothione Reductase From Mycobacterium Tuberculosis In Complex With Respiri-1093
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: OXIDOREDUCTASE Ligands: FAD, A1H3R |
 |
Neutron Structure Of Ferredoxin-Nadp+ Reductase From Maize Root -Reduced Form
Organism: Zea mays
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, FDA |
 |
Neutron Structure Of Ferredoxin-Nadp+ Reductase From Maize Root -Oxidized Form
Organism: Zea mays
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, MES |
 |
High Resolution Structure Of Ferredoxin-Nadp+ Reductase From Maize Root - Oxidized Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, GOL, FAD |
 |
High Resolution Structure Of Ferredoxin-Nadp+ Reductase From Maize Root - Reduced Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, FDA |
 |
R115A Mutant Of Ferredoxin-Nadp+ Reductase From Maize Root - Oxidized Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, MES |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN Ligands: FAD |
 |
Structure Of Mycobacterial Ndh2 (Type Ii Nadh:Quinone Oxidoreductase) With 2-Mercapto-Quinazolinone Inhibitor.
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN Ligands: FAD, A1BNR |
 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm679 (Ethyl 1,4-Dimethyl-5-((6-(Trifluoromethyl)Pyridin-3-Yl)Methyl)-1H-Pyrazole-3-Carboxylate)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: A1A51, FMN, OG6, CIT, LDA |
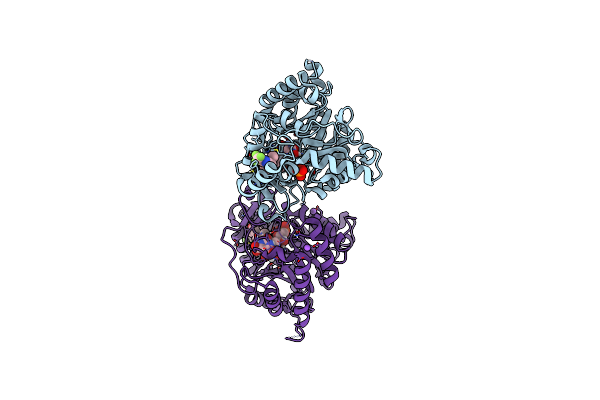 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm681 (N-Cyclopropyl-1,4-Dimethyl-5-((6-(Trifluoromethyl)Pyridin-3-Yl)Methyl)-1H-Pyrazole-3-Carboxamide)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: A1A52, FMN, OG6, NA |
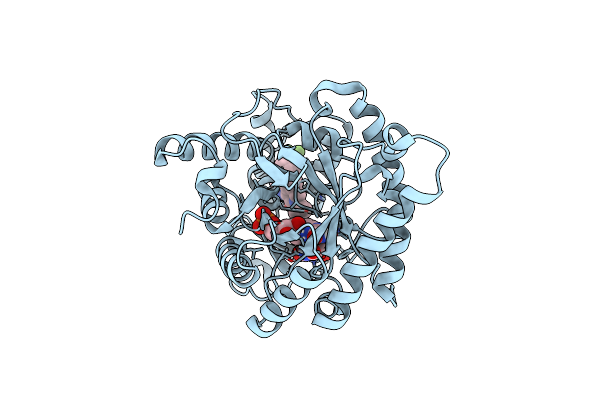 |
Crystal Structure Of Plasmodium Falciparum Dihydroorotate Dehydrogenase Bound With Inhibitor Dsm959 (6-Cyclopropyl-2,4-Dimethyl-3-((6-(Trifluoromethyl)Pyridin-3-Yl)Methyl)-2,6-Dihydro-7H-Pyrazolo[3,4-C]Pyridin-7-One)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-01-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: A1A53, FMN, OG6 |
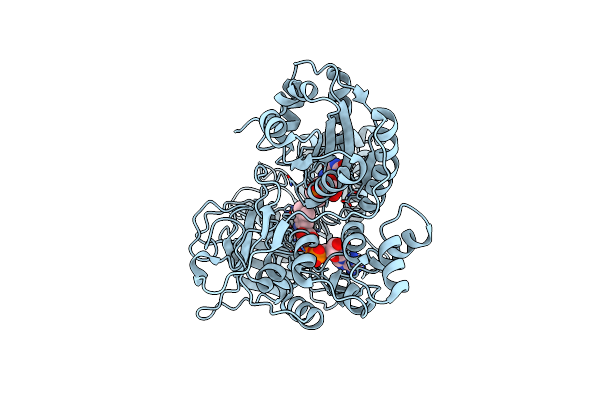 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: OXIDOREDUCTASE Ligands: FMN, FAD |
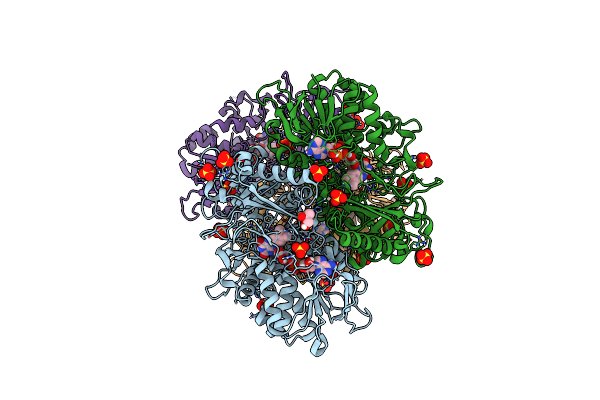 |
Organism: Hebeloma cylindrosporum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, PGO |
 |
L-Amino Acid Oxidase 4 (Hclaao4) From The Fungus Hebeloma Cylindrosporum In Complex With L-Glutamine
Organism: Hebeloma cylindrosporum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: FDA, GLN, SO4, PGR, PGO |
 |
L-Amino Acid Oxidase 4 (Hclaao4) From The Fungus Hebeloma Cylindrosporum In Complex With L-Glutamate
Organism: Hebeloma cylindrosporum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: FDA, GLU, SO4, PGR, PGO |
 |
L-Amino Acid Oxidase 4 (Hclaao4) From The Fungus Hebeloma Cylindrosporum In Complex With L-Phenylalanine
Organism: Hebeloma cylindrosporum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: FDA, PHE, SO4, PGR |
 |
L-Amino Acid Oxidase 4 (Hclaao4) From The Fungus Hebeloma Cylindrosporum In Complex With N-Epsilon-Acetyl-L-Lysine
Organism: Hebeloma cylindrosporum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-08-14 Classification: OXIDOREDUCTASE Ligands: FDA, ALY, SO4, PGR, PGO |
 |
Crystal Structure Of Ancestral L-Galactono-1,4-Lactone Dehydrogenase: In Complex With L-Galactono-1,4-Lactone
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-05-01 Classification: FLAVOPROTEIN Ligands: FAD, X8X |
 |
Crystal Structure Of Ancestral L-Galactono-1,4-Lactone Dehydrogenase: G413N Variant In Complex With L-Gulono-1,4-Lactone
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-05-01 Classification: FLAVOPROTEIN Ligands: FAD, X8L |

