Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of C43S Terpredoxin, A [2Fe-2S] Ferredoxin From Pseudomonas Sp
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
 |
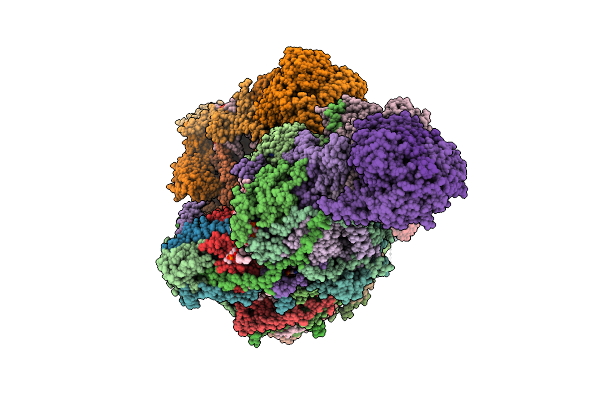
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: CDL, PEE, NDP, ZMP, ADP, 3PE, PC1, PLX, XEW, SF4, FES, MG, ZN, FMN |
 |
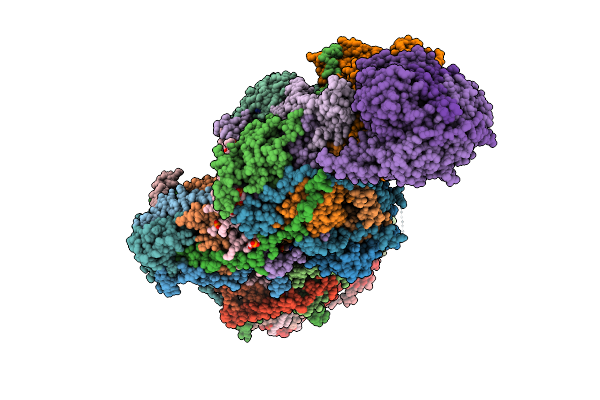
Organism: Paracoccus denitrificans pd1222
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: ELECTRON TRANSPORT Ligands: PC1, U10, HEM, CA, HEC, 3PE, DU0, FES, 3PH, CDL, HEA, CU, MN, CUA, ZN |
 |
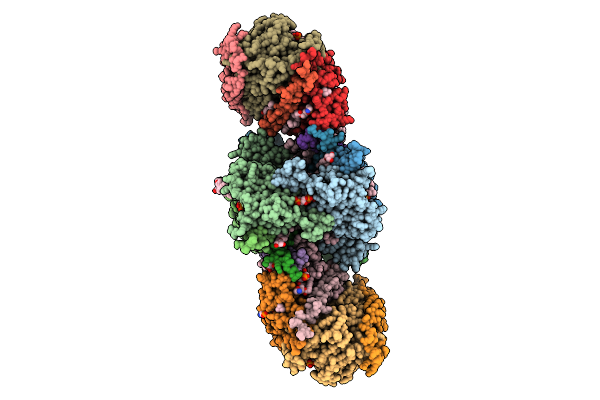
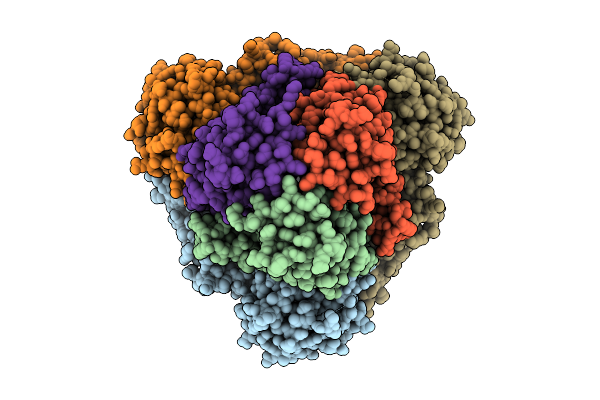
Cryo-Em Structure Of Spinacia Oleracea Cytochrome B6F Complex With Bound Plastocyanin
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: HEM, HEC, UMQ, PL9, CLA, FES, SQD, BCR, CU |
 |
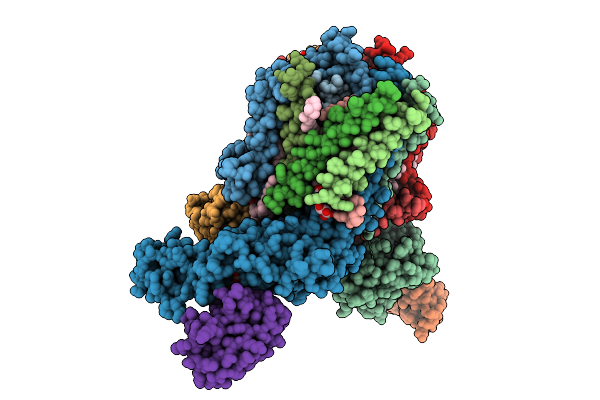
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: FE2, FES, HCI |
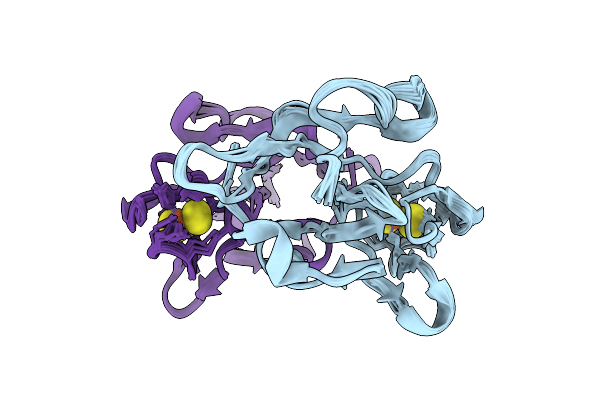 |
Soluble Domain Of Kustd1480, A Rieske Iron-Sulfur Cluster Protein From Kuenenia Stuttgartiensis
Organism: Candidatus kuenenia sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ELECTRON TRANSPORT Ligands: FES |
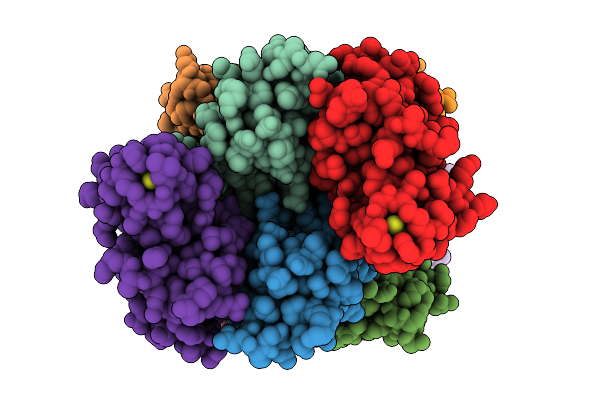 |
Soluble Domain Of Kuste4569, A Rieske Iron-Sulfur Cluster Protein From Kuenenia Stuttgartiensis
Organism: Candidatus kuenenia sp.
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ELECTRON TRANSPORT Ligands: FES, SO4 |
 |
 |
 |
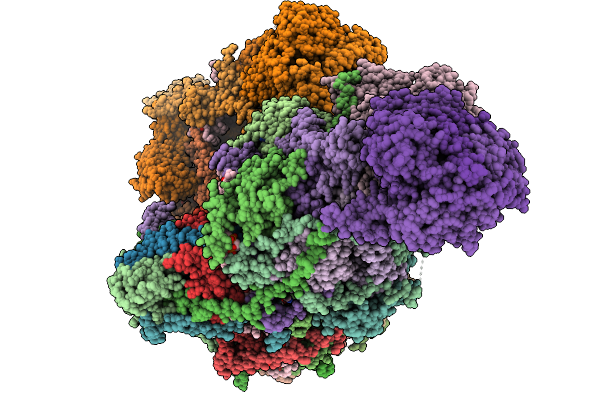
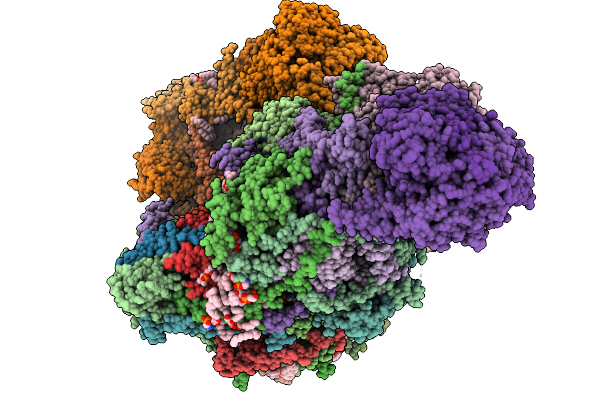
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10, Alternative Orientation (Sc-Metc1-Ii)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, PX2, U10, SF4, FES, MG, MF8, ZN, FMN |
 |
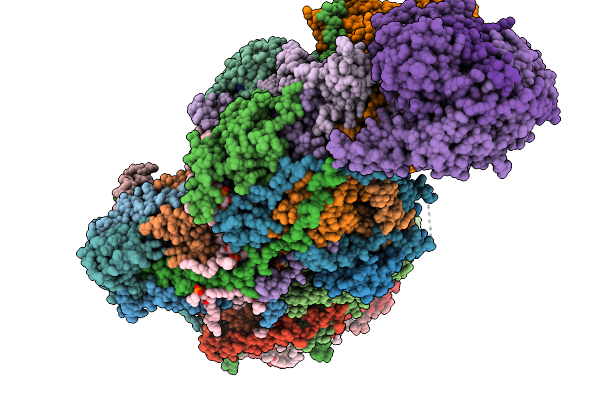
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10 (Sc-Metc1-I)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, U10, SF4, FES, MG, MF8, ZN, FMN |
 |
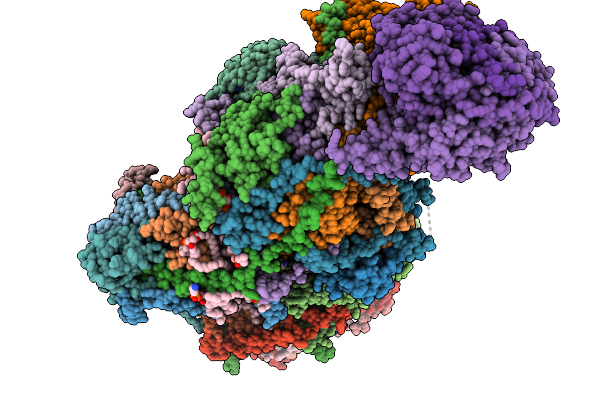
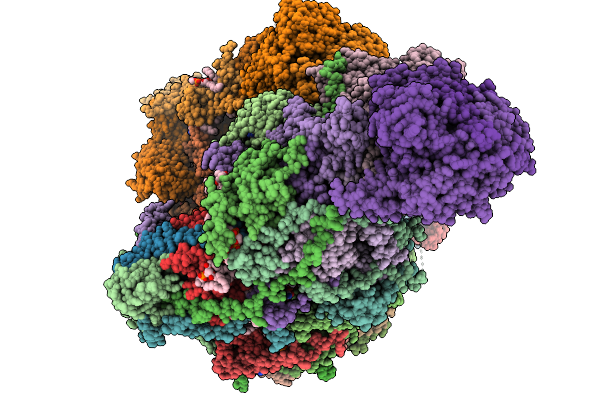
Complex I From Respirasome Closed State 1 Bound By Metformin (Sc-Metc1-Iii)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, SF4, FES, MG, MF8, ZN, FMN |
 |
 |
 |
 |
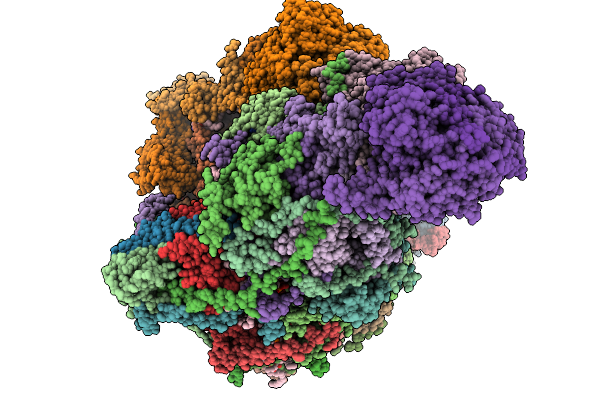
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, NDP, PEE, ZMP, ADP, PLX, 3PE, XEW, 6F6, SF4, FES, MG, ZN, FMN |
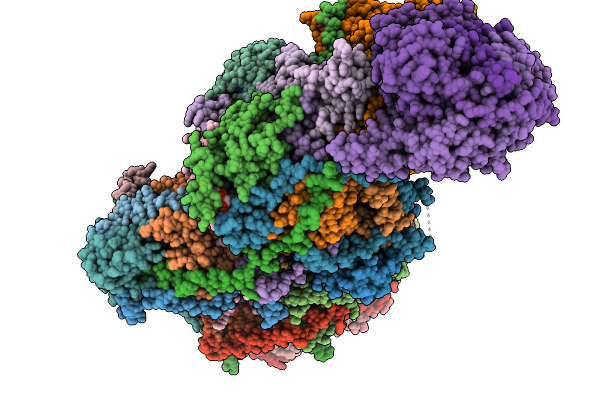 |
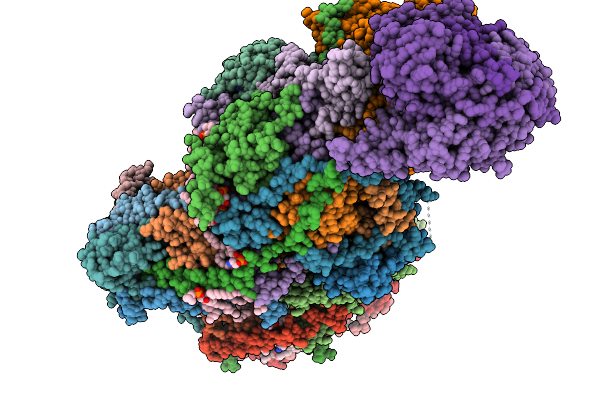
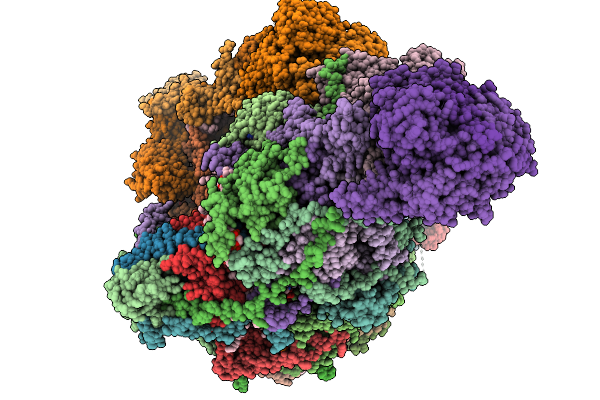
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10 (Sc-Metc1-Iv)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, NDP, PEE, PC1, ZMP, ADP, PLX, 3PE, U10, SF4, FES, MG, MF8, ZN, FMN |
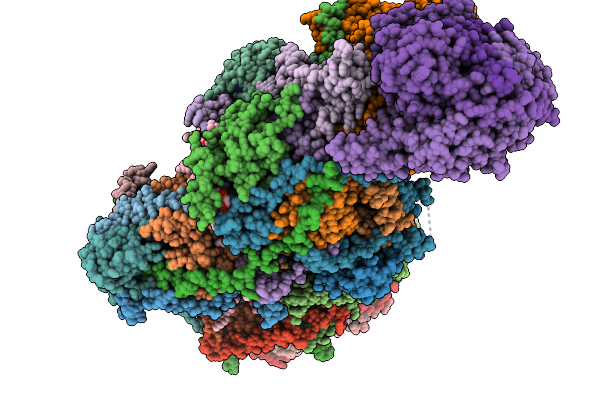 |
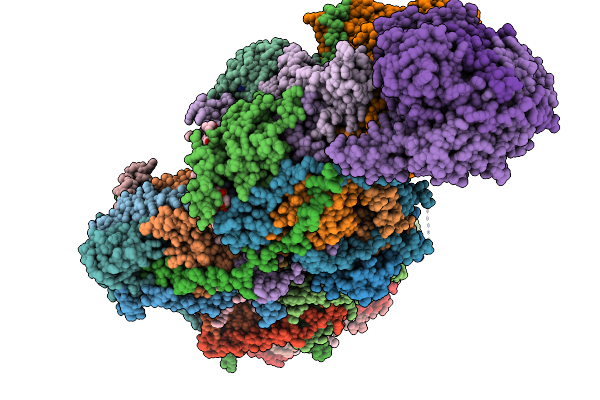
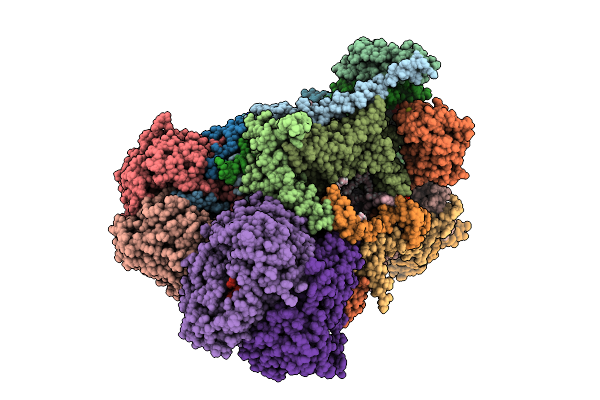
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10 (Sc-Metc1-V)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, NDP, PEE, ZMP, ADP, PLX, 3PE, PC1, SF4, FES, MG, ZN, U10, MF8, FMN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: CDL, 3PE, FES, PLX, HEC, HEM, PEE, U10, PC1 |