Search Count: 60
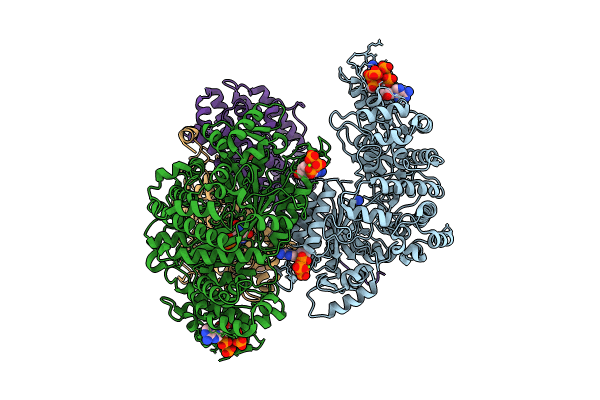 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: OXIDOREDUCTASE Ligands: DTP, MG, ATP, UNL, A1A3L, FEO |
 |
Crystal Structure Of A Flavodiiron Protein S262Y Mutant In The Oxidized State From Escherichia Coli
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, FEO, GOL |
 |
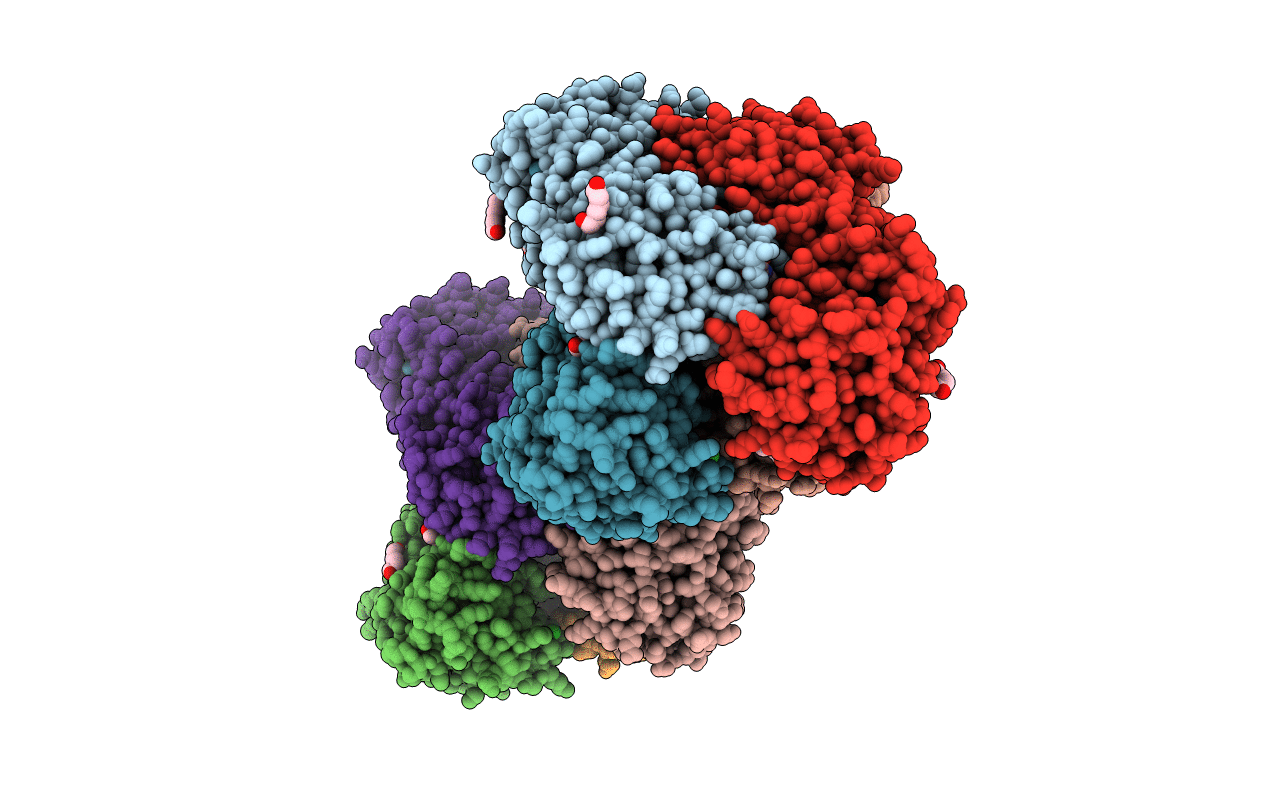
Structure Of The Neisseria Gonorrhoeae Ribonucleotide Reductase In The Inactive State
Organism: Neisseria gonorrhoeae
Method: ELECTRON MICROSCOPY Release Date: 2022-01-05 Classification: OXIDOREDUCTASE Ligands: CDP, DTP, MG, FEO |
 |
Aerobic Crystal Structure Of F420H2-Oxidase From Methanothermococcus Thermolithotrophicus At 1.8A Resolution Under 125 Bars Of Krypton
Organism: Methanothermococcus thermolithotrophicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: OXIDOREDUCTASE Ligands: HEZ, FEO, KR, FMN, CL |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2020-04-08 Classification: OXIDOREDUCTASE Ligands: TTP, MG, GDP, FEO |
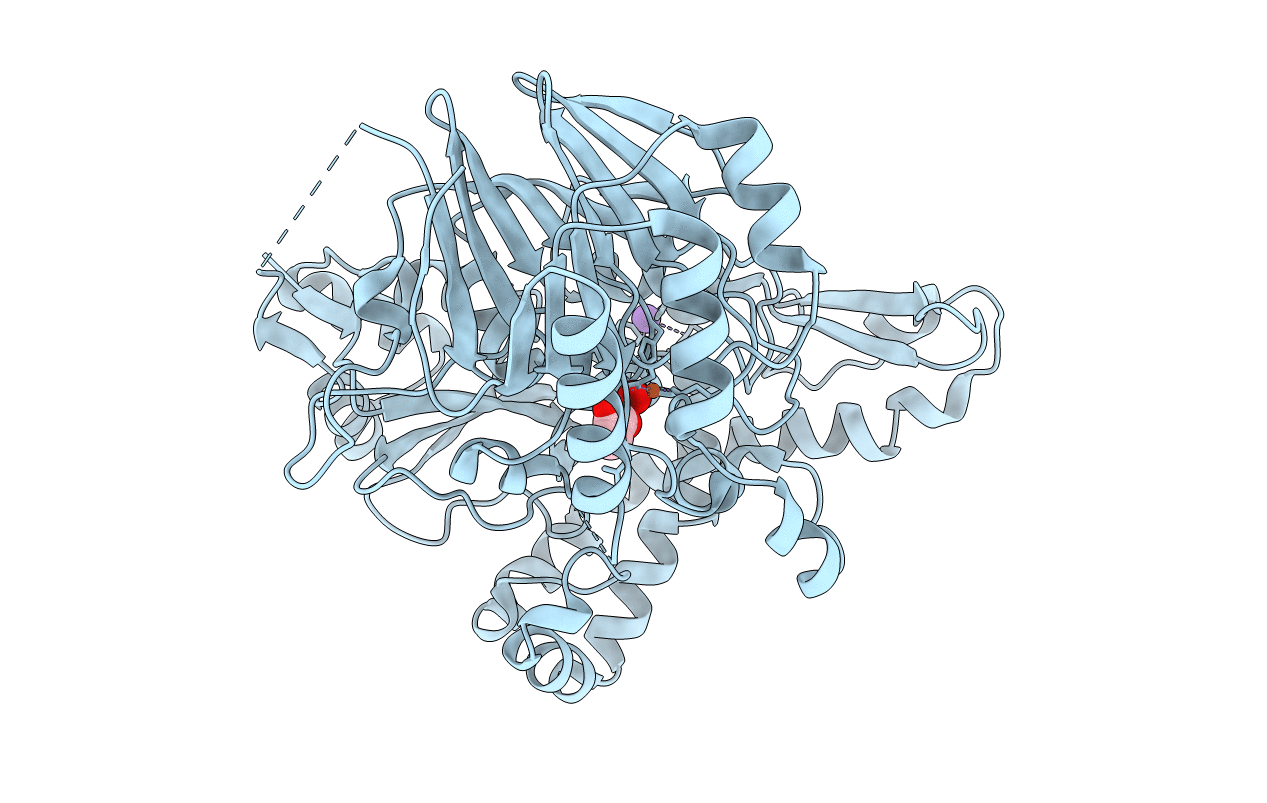 |
Organism: Streptomyces venezuelae (strain atcc 10712 / cbs 650.69 / dsm 40230 / jcm 4526 / nbrc 13096 / pd 04745)
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2017-04-26 Classification: OXIDOREDUCTASE Ligands: FEO, K, ACT |
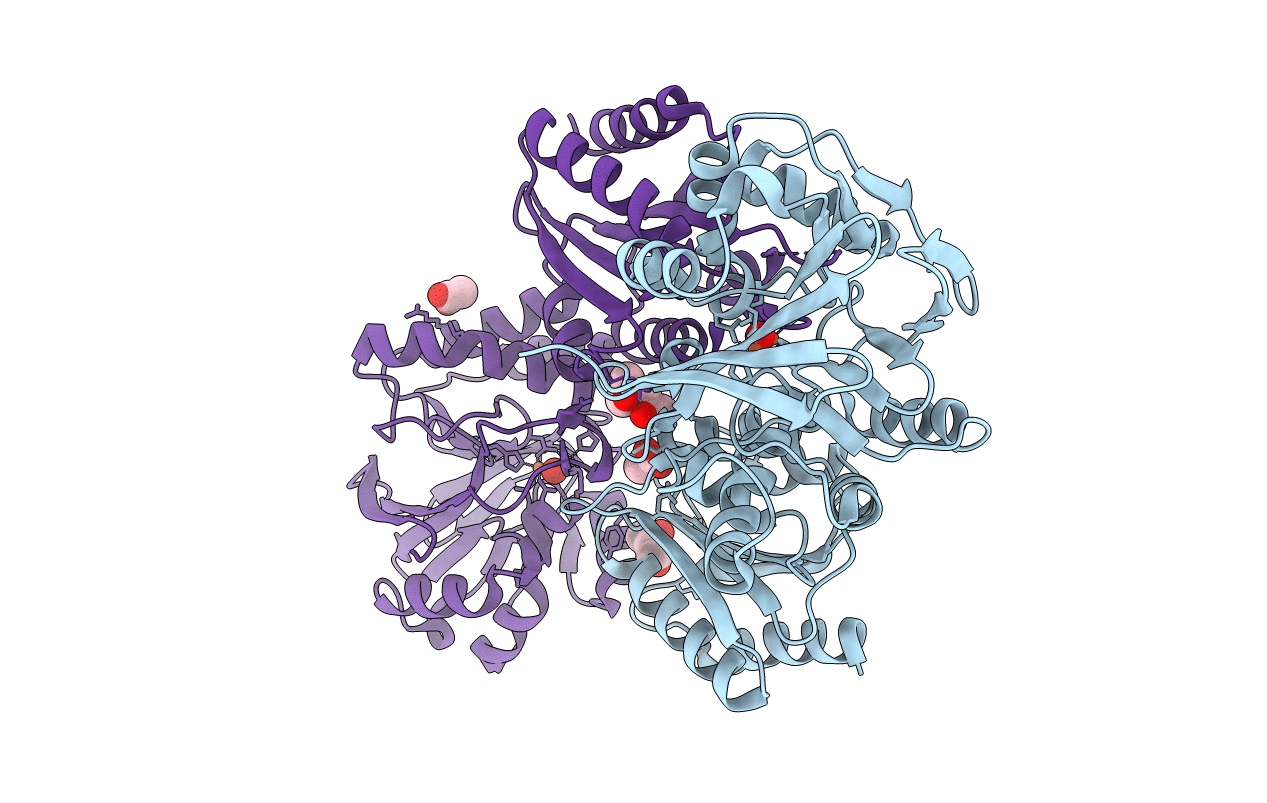 |
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2017-04-19 Classification: ELECTRON TRANSPORT Ligands: FEO, CL, ACT, MRD |
 |
Oxidized Flavodiiron Core Of Escherichia Coli Flavorubredoxin, Including The Fe-4Sg Atoms From Its Rubredoxin Domain
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-10-19 Classification: OXIDOREDUCTASE Ligands: FEO, FMN, PO4, CAC, ACY, GOL, FE |
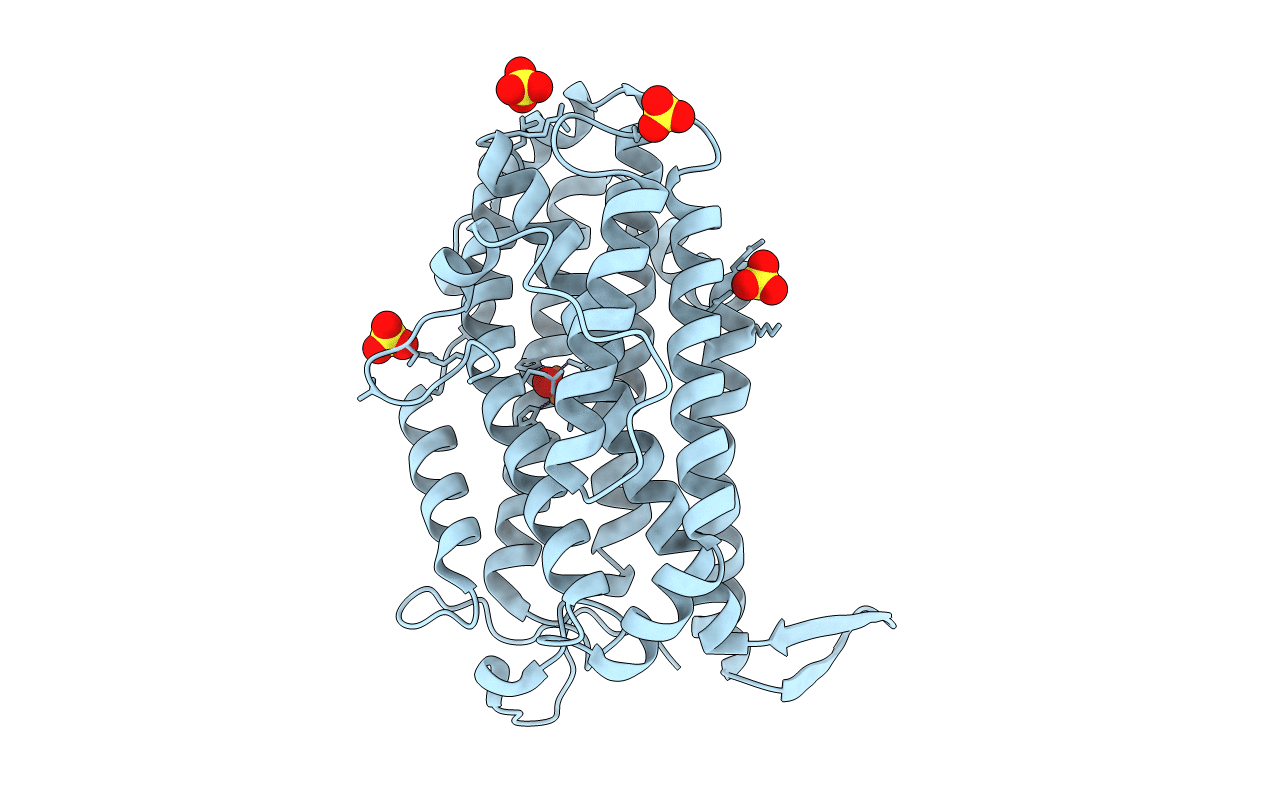 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-07-13 Classification: OXIDOREDUCTASE Ligands: FEO, SO4 |
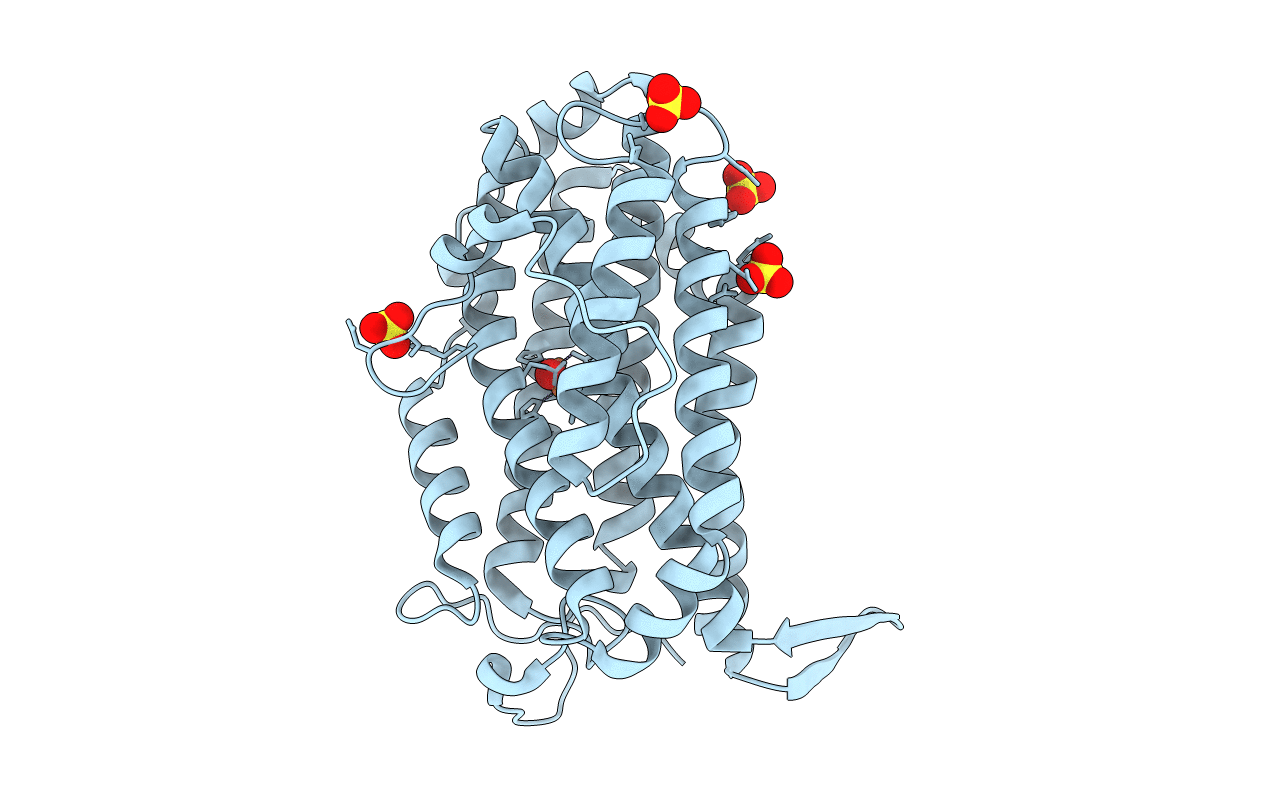 |
Organism: Escherichia coli 1303
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE Ligands: FEO, SO4 |
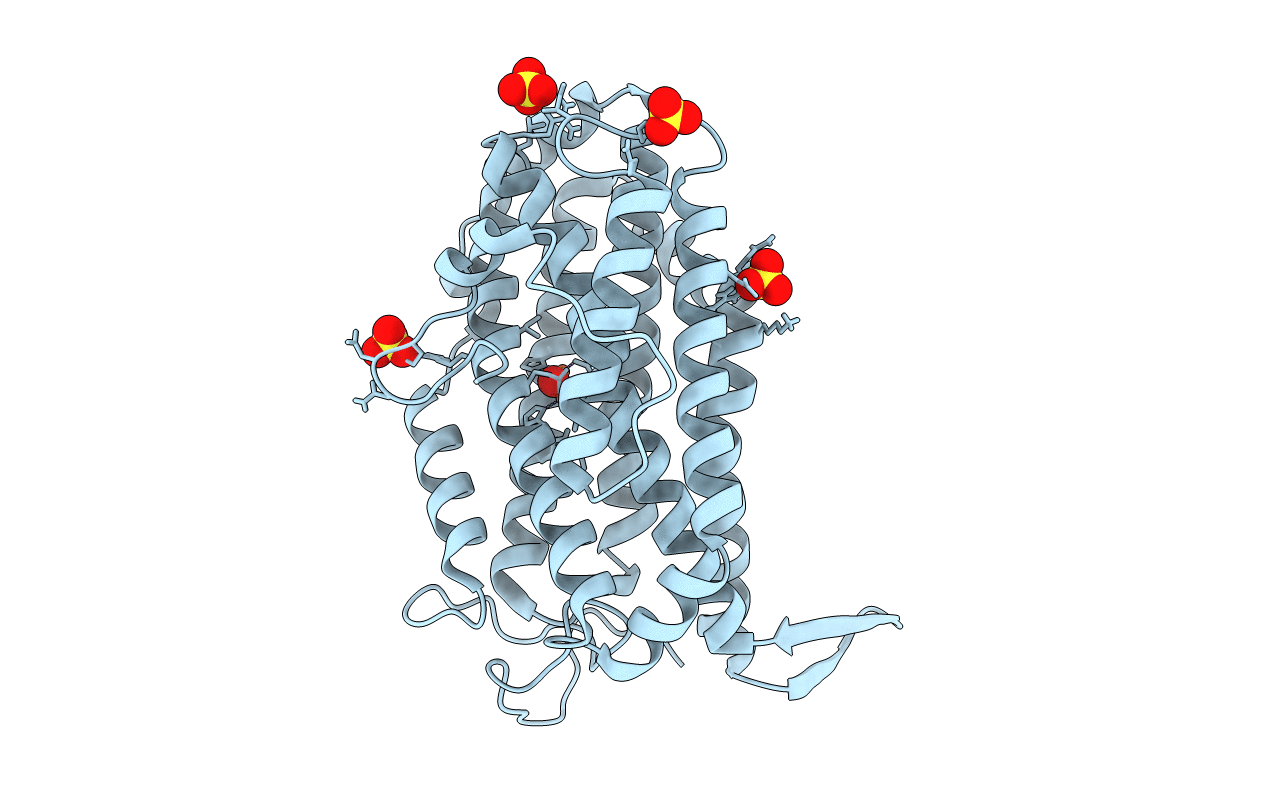 |
Organism: Escherichia coli 1303
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE Ligands: FEO, SO4, CL |
 |
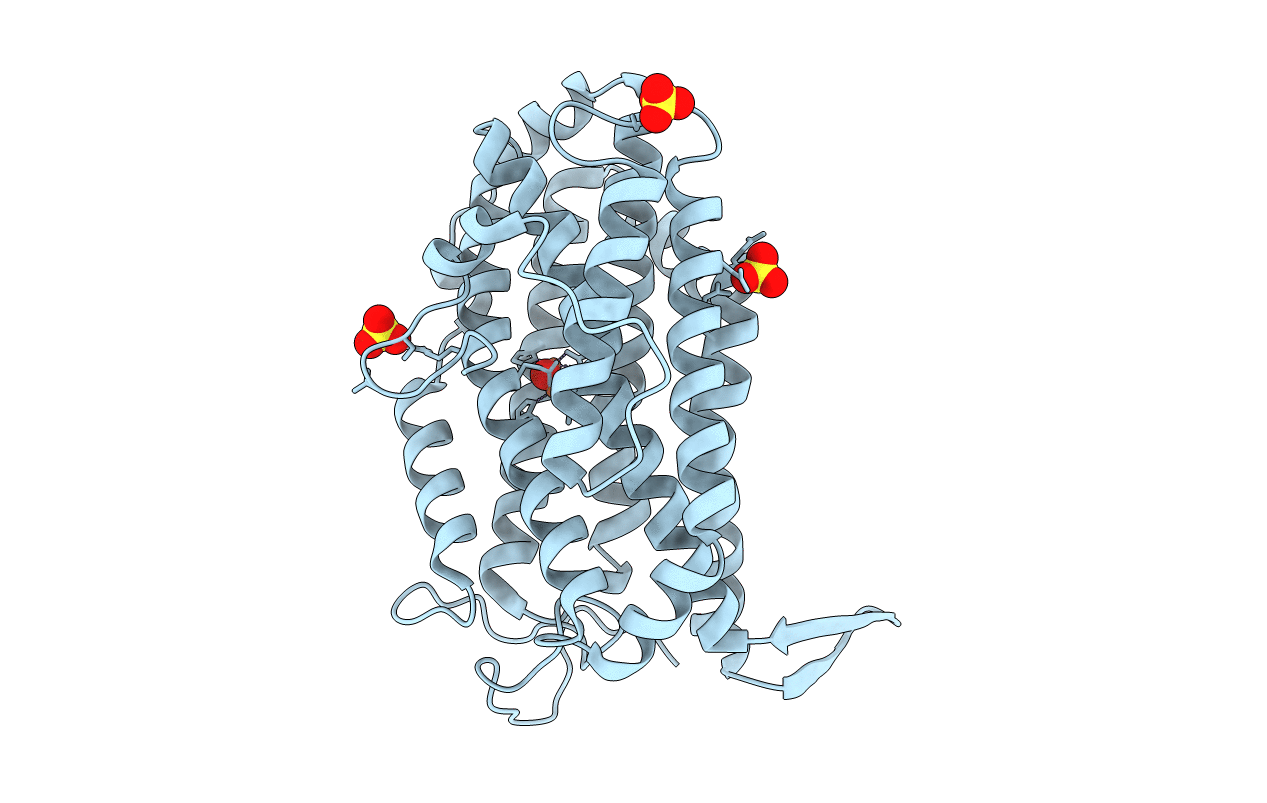
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE Ligands: FEO, SO4 |
 |
Organism: Escherichia coli 1303
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE Ligands: FEO, SO4 |
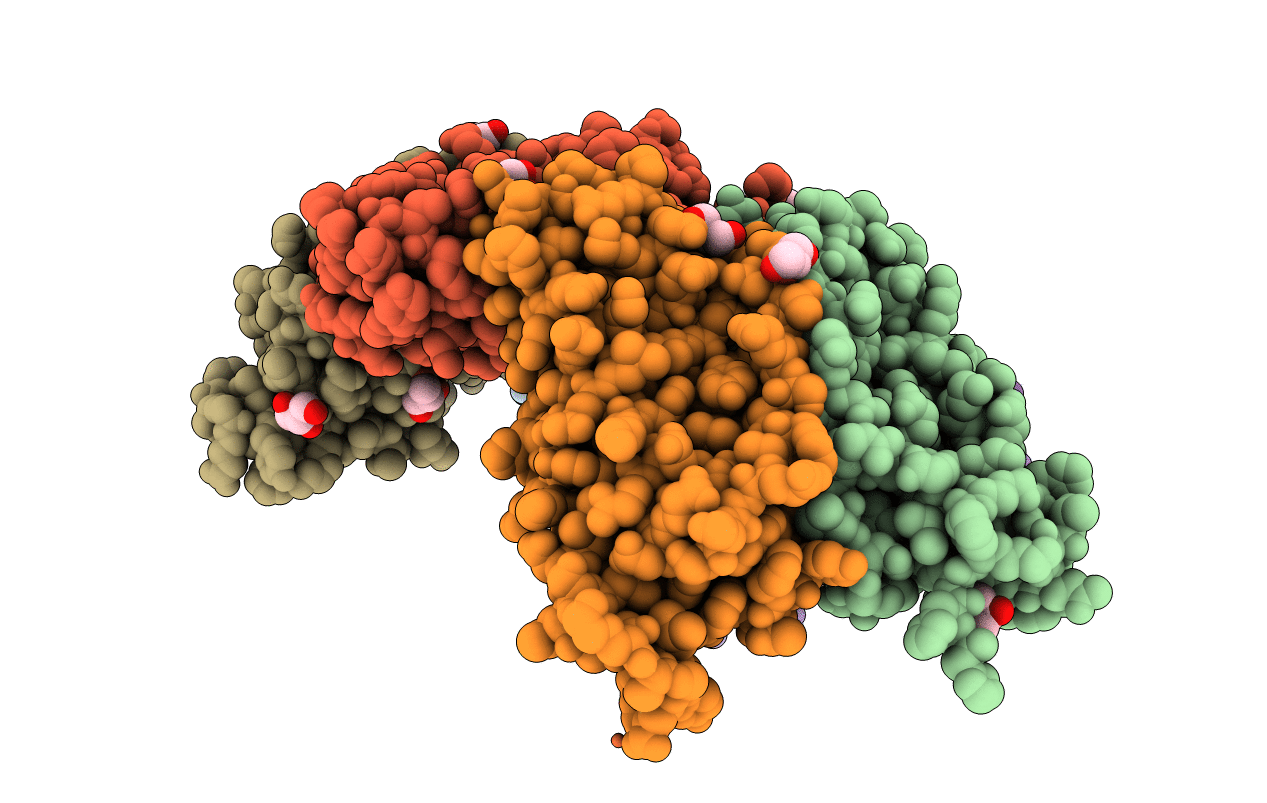 |
Crystal Structures Of Ferritin Mutants Reveal Diferric-Peroxo Intermediates
Organism: Escherichia coli dh1
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2016-06-15 Classification: OXIDOREDUCTASE Ligands: FEO, OH, GOL, FE, FE2, OXY, PER, O |
 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex Bound To Cdp And Datp At 2.97 Angstroms Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: CDP, DAT, MG, DTP, FEO |
 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex Bound To Udp And Datp At 3.25 Angstroms Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: UDP, DTP, MG, FEO |
 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex Bound To Adp And Dgtp At 3.40 Angstroms Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: DGT, MG, ADP, DAT, FEO |
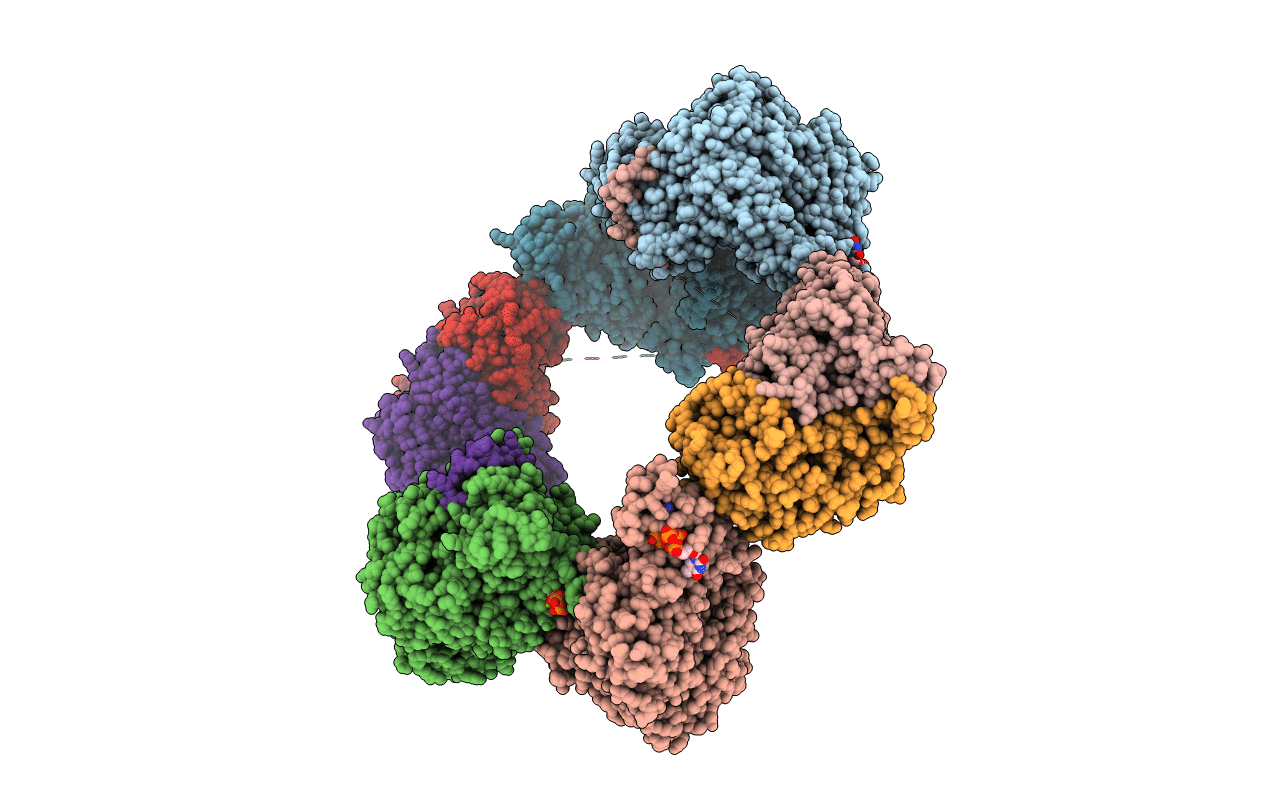 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex Bound To Gdp And Ttp At 3.20 Angstroms Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: GDP, DAT, MG, TTP, FEO |
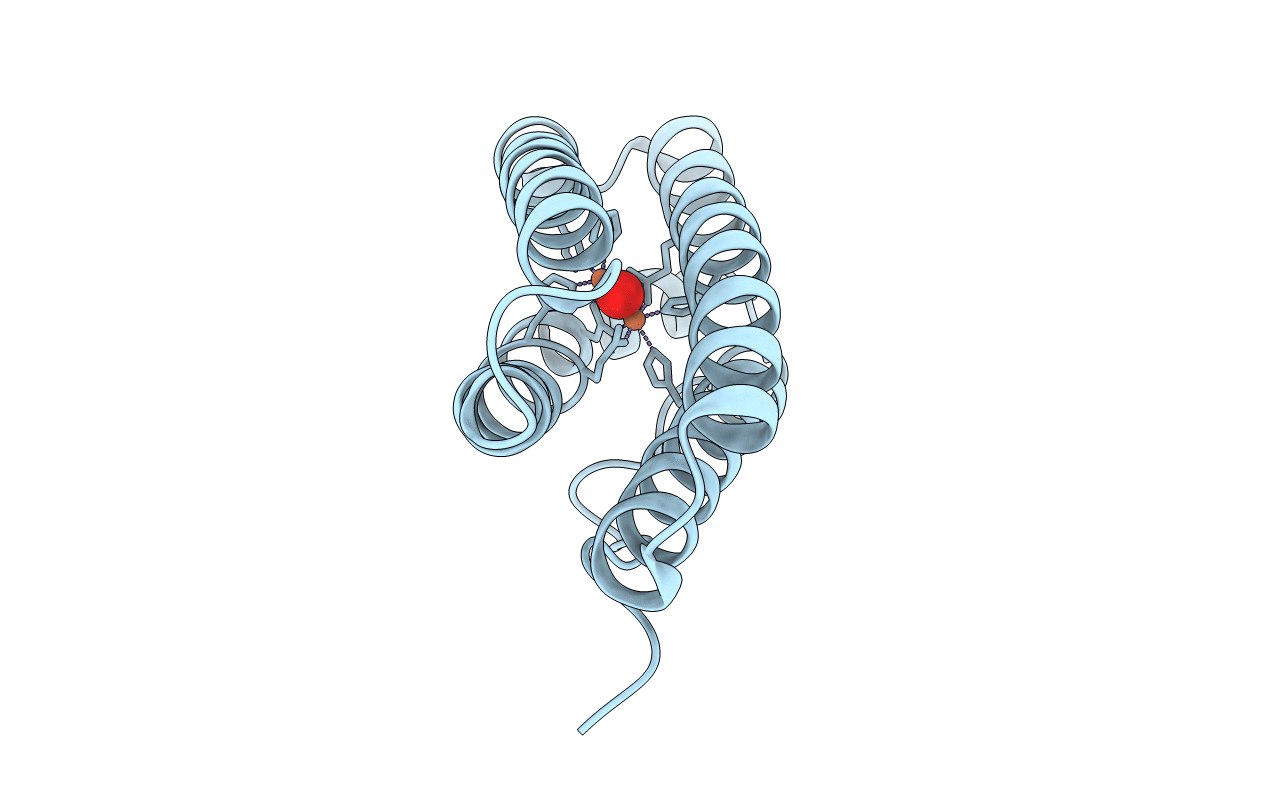 |
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-03-19 Classification: METAL BINDING PROTEIN Ligands: FEO |
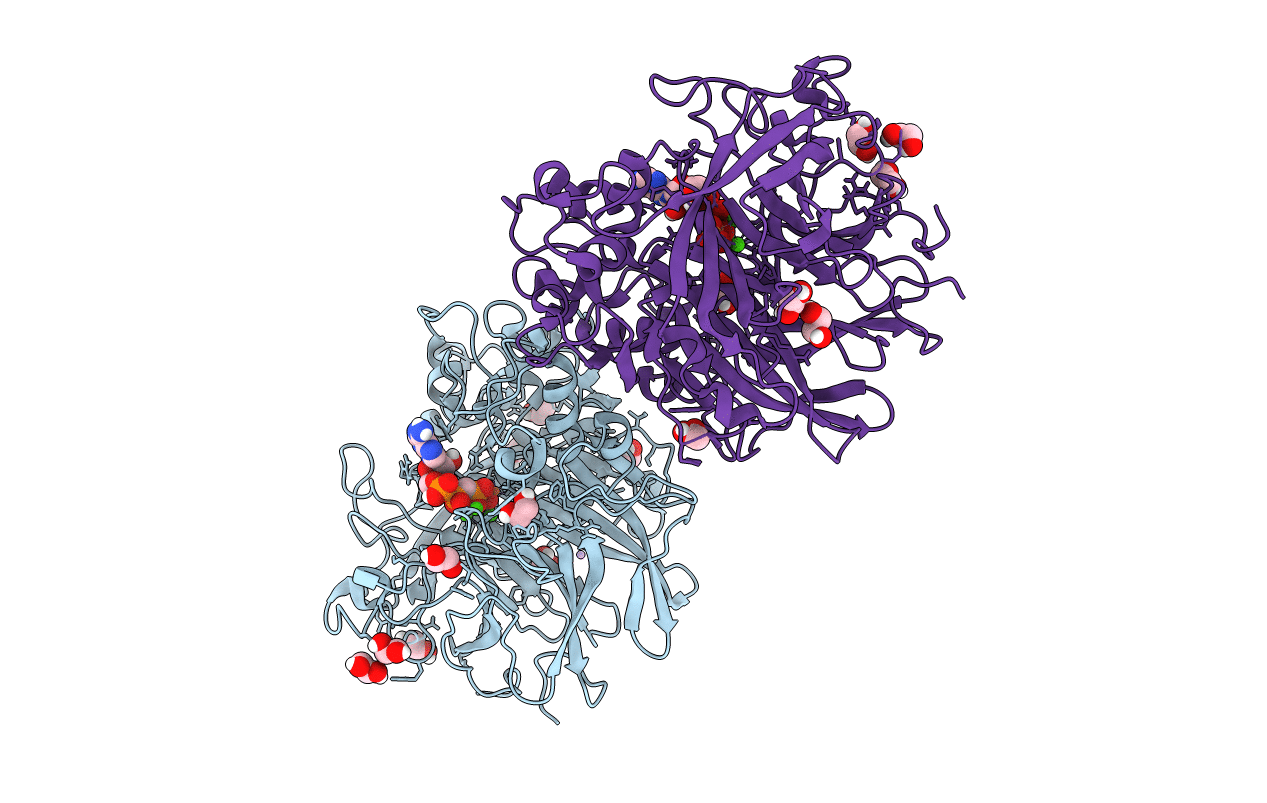 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2013-03-20 Classification: HYDROLASE Ligands: ACP, CL, LI, CA, FEO, EDO |

