Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
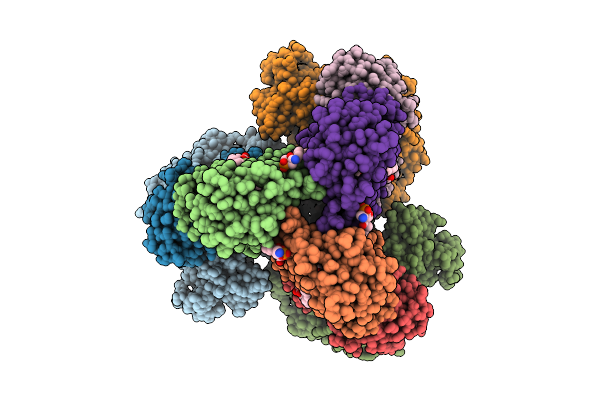 |
Cryo-Em Structure Of The Apo-Form Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SIN, SF4, FES, F3S, HEM, LMT, PGV, PEV |
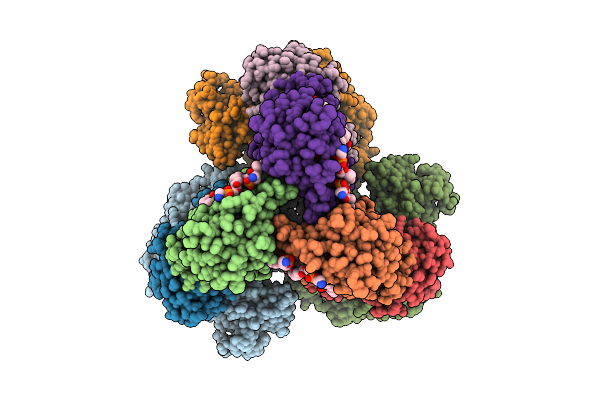 |
Cryo-Em Structure Of The Lipid-Bound Succiante Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SIN, SF4, FES, F3S, HEM, LMT, PGV, PEV |
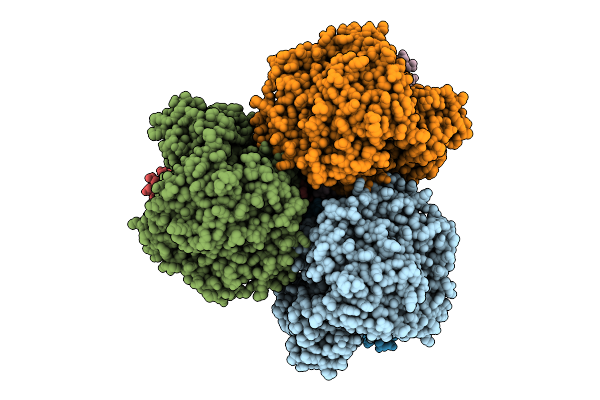 |
Cryo-Em Structure Of The Mk7-Bound Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SF4, FES, F3S, HEM, LMT, MQ7 |
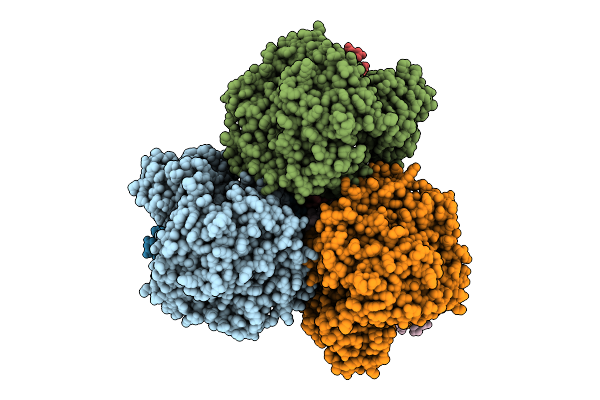 |
Cryo-Em Structure Of The Mk4-Bound Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SF4, FES, F3S, HEM, LMT, 1L3 |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, PEE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Uq1-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, UQ1, PEE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Pydiflumetofen-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, A1EE4, PEE |
 |
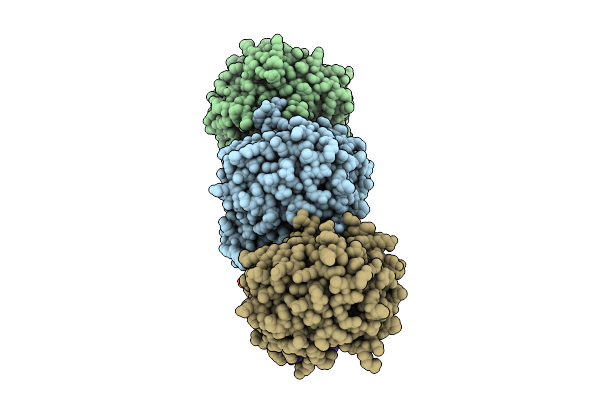
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Y19315-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, A1EKV, PEE |
 |
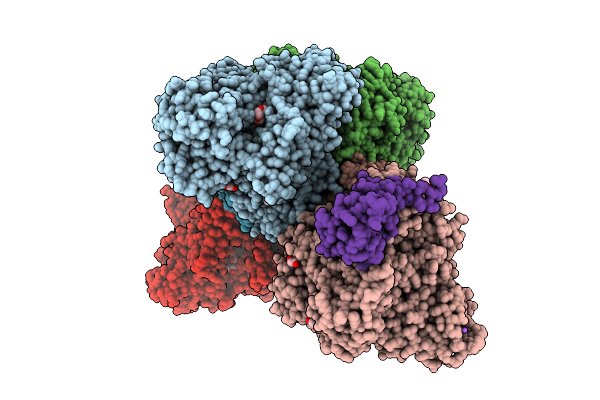
Structure Of The Complex Between Human Lias And H-Protein In The Presence Of S-Adenosyl-L-Methionine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, F3S, SAM, YVI, PO4 |
 |
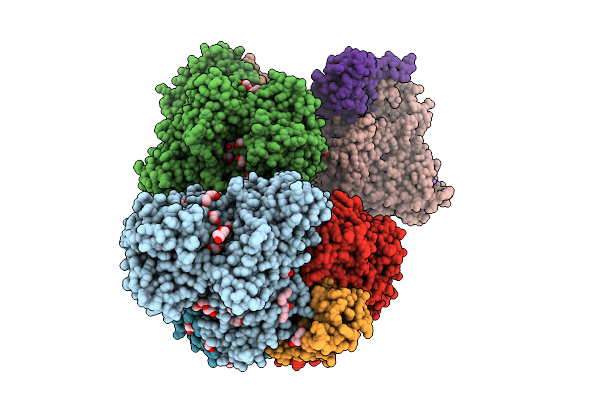
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: MGD, 4MO, F3S, PEG, EDO, 1PE, PGE, GOL, O, NA, FES, P4G, PG0 |
 |
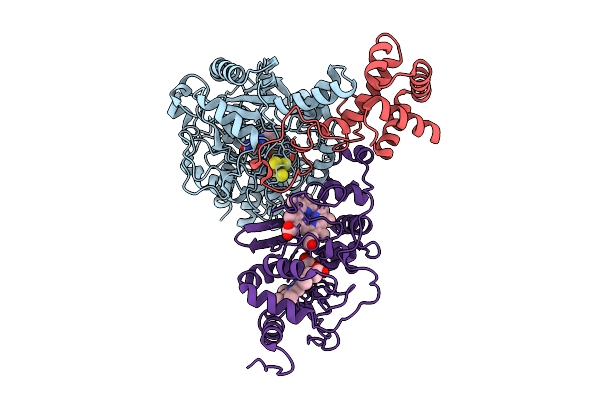
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: MO, MGD, F3S, EDO, PEG, SBO, P33, PGE, O, NA, FES, GOL, 4MO, P4G, 1PE, PG0 |
 |
Cryo-Em Structure Of Fructose Dehydrogenase Variant From Gluconobacter Japonicus Truncating Heme 1C And C-Terminal Hydrophobic Regions
Organism: Gluconobacter japonicus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: FAD, F3S, HEC |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: SF4, FES, F3S, FE, NI, CMO, CO2, TRS |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: SF4, FES, F3S, FE, NI, CO2, CMO, TRS |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: SF4, FES, F3S, FE, NI, CMO, CO2, TRS, PEG |
 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: SF4, FES, F3S, FE, NI, CO2, CMO, TRS |
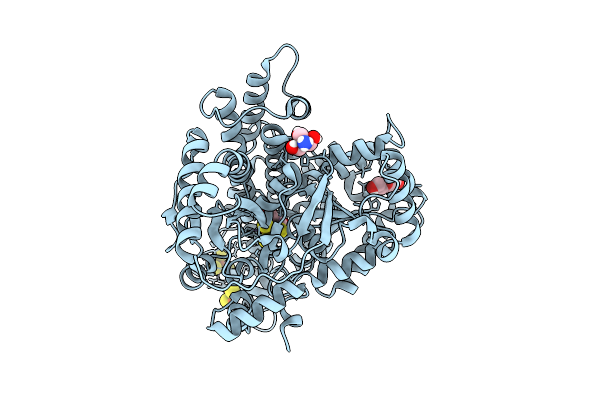 |
Organism: Carboxydothermus hydrogenoformans z-2901
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: SF4, FES, F3S, FE, NI, CO2, CMO, TRS, PEG |
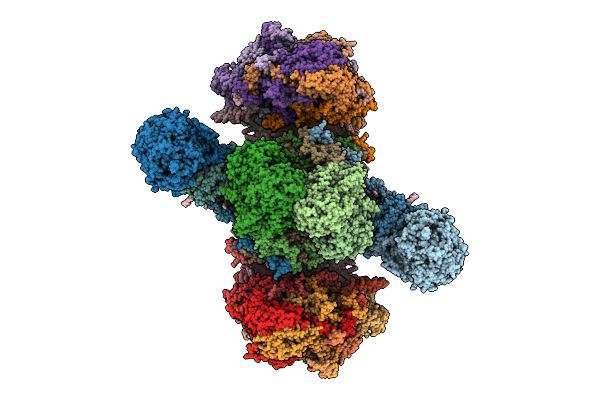 |
Perkinsus Marinus Respiratory Supercomplex Cii2Ciii2Civ2 In An Intermediate State
|
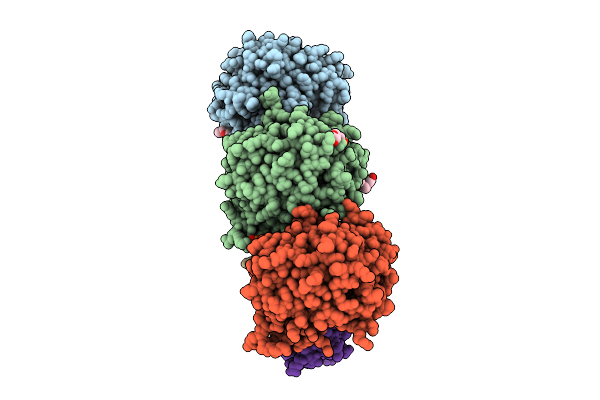 |
Structure Of The Complex Between Human Lias And H-Protein In The Presence Of 5'-Deoxyadenosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: BIOSYNTHETIC PROTEIN Ligands: 5AD, SF4, F3S, PEG, PG4, MET, YVI, 2PE, PGE, PO4 |
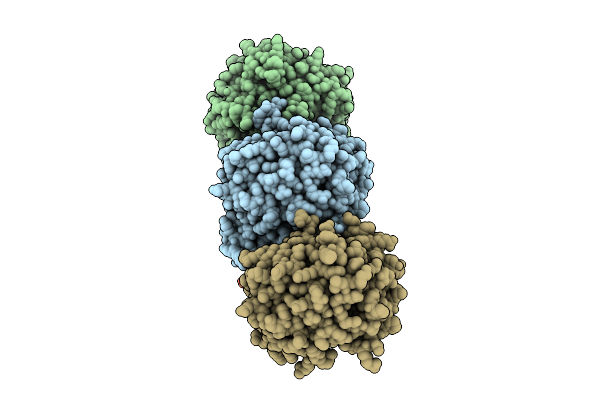 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: BIOSYNTHETIC PROTEIN Ligands: SF4, F3S, SAM, YVI, PO4 |