Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
A Double Mutant Of Serratia Marcescens Hemophore Receptor Hasr In Complex With Its Hemophore Hasa And Heme
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-06-29 Classification: TRANSPORT PROTEIN Ligands: HEM |
 |
Structure Of The P53 Cancer Mutant Y220C With Bound Small Molecule 7- Ethyl-3-(Piperidin-4-Yl)-1H-Indole
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2015-12-16 Classification: TRANSCRIPTION Ligands: ZN, PEG, 92O |
 |
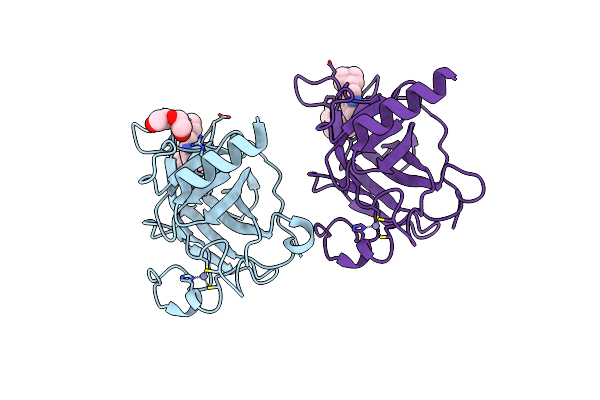
Structure Of The P53 Cancer Mutant Y220C With Bound Small-Molecule Stabilizer Phikan5149
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2015-12-16 Classification: TRANSCRIPTION Ligands: ZN, UL7 |
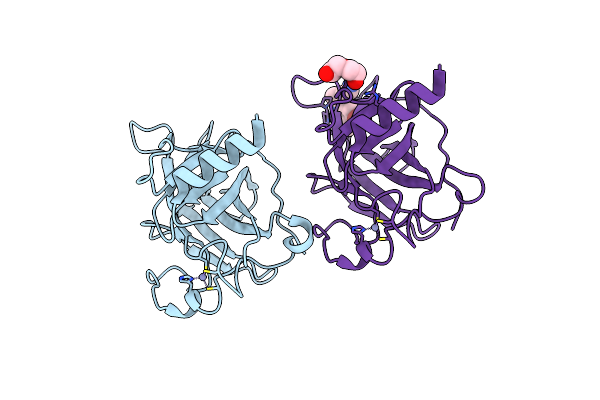 |
Structure Of The P53 Cancer Mutant Y220C In Complex With An Indole- Based Small Molecule
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2015-12-16 Classification: SIGNALING PROTEIN Ligands: ZN, PEG, RZH |
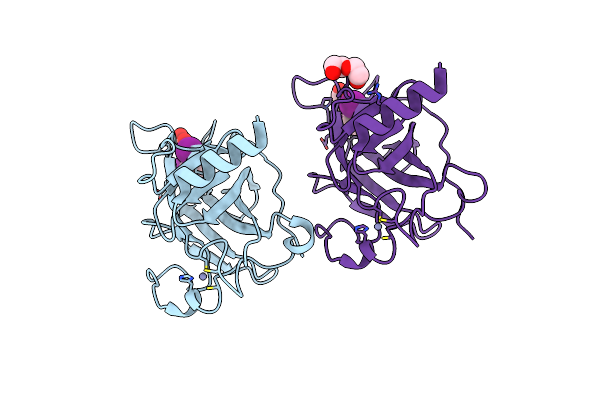 |
Structure Of The P53 Cancer Mutant Y220C In Complex With 2-Hydroxy-3, 5-Diiodo-4-(1H-Pyrrol-1-Yl)Benzoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2015-12-16 Classification: CELL CYCLE Ligands: Y0V, ZN, PEG |
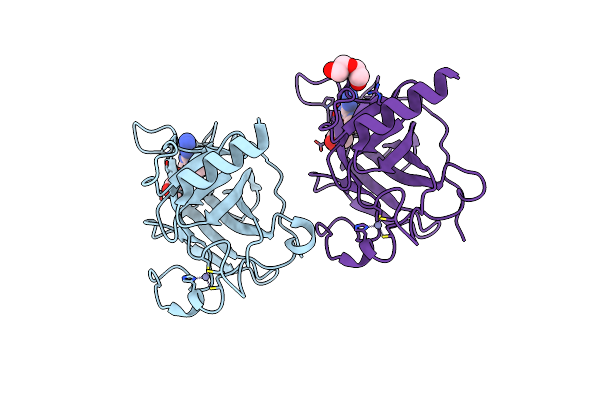 |
Structure Of The P53 Cancer Mutant Y220C With Bound Small Molecule Phikan7099
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2015-12-16 Classification: CELL CYCLE Ligands: ZN, GOL, GOH, PEG |
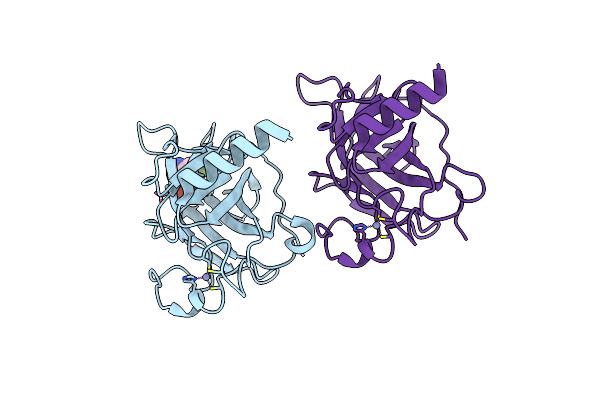 |
Structure Of The P53 Cancer Mutant Y220C With Bound 3-Bromo-5-(Trifluoromethyl)Benzene-1,2-Diamine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-12-16 Classification: CELL CYCLE Ligands: UFV, ZN |
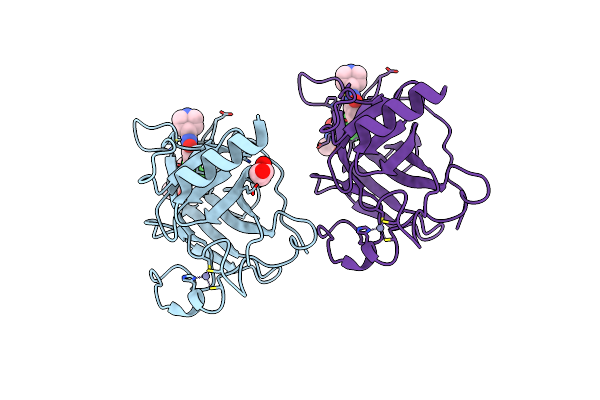 |
Structure Of The P53 Cancer Mutant Y220C With Bound Small Molecule Phikan883
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2015-12-16 Classification: CELL CYCLE Ligands: FY8, GOL, ZN |
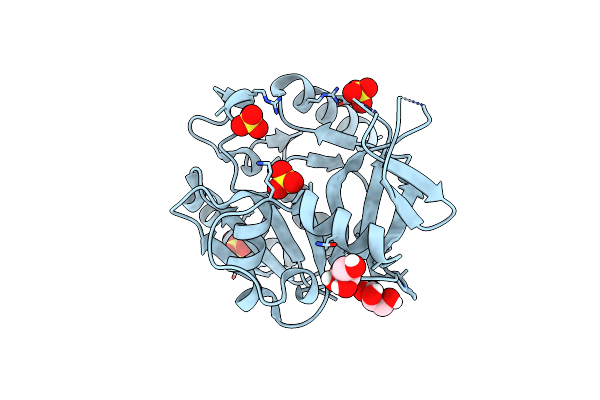 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-18 Classification: TRANSFERASE Ligands: GOL, SO4 |
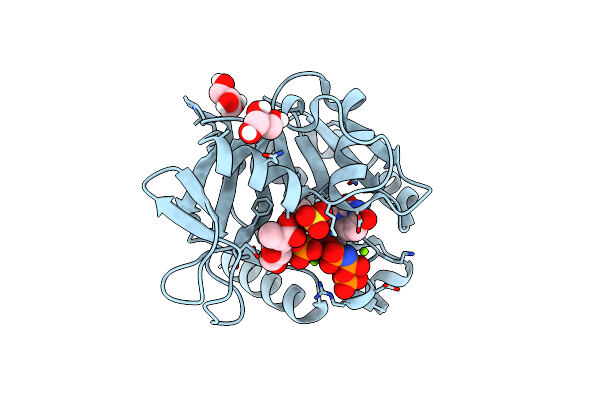 |
Organism: Pseudomonas putida (strain bird-1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-03-18 Classification: TRANSFERASE Ligands: GOL, SO4, MG, 2KH, 491 |
 |
Organism: Pseudomonas putida (strain bird-1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-18 Classification: TRANSFERASE Ligands: GOL, SO4, 491, 2PN |
 |
Crystal Structure Of The Lecb Lectin From Pseudomonas Aeruginosa In Complex With Methyl 6-(2,4,6-Trimethylphenylsulfonylamido)-6-Deoxy-Alpha-D-Mannopyranoside
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2013-09-18 Classification: SUGAR BINDING PROTEIN Ligands: CA, MMA, F1A, GOL, EDO, SO4 |
 |
Crystal Structure Of The Large Fragment Of Dna Polymerase I From Thermus Aquaticus In A Closed Ternary Complex With Trapped 4'-Ethylated Dttp
Organism: Thermus aquaticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-07-07 Classification: Transferase/DNA Ligands: HXZ, MG, 15P, GOL, ACT |
 |
Crystal Structure Of The Large Fragment Of Dna Polymerase I From Thermus Aquaticus In A Closed Ternary Complex With Trapped 4'-Methylated Dttp
Organism: Thermus aquaticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-07-07 Classification: Transferase/DNA Ligands: HXB, MG, GOL, PEG, 15P, ACT |