Search Count: 56
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z943693514
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, DMS, BR, NA, GT4 |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z2856434836
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, BR, LPZ |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z2204875953
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, Q7L, BR |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z2856434890
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, NZD, BR |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z32399802
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, BR, A1I3U |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z2017861827
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, A1BE9, BR |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z319545618
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, RXS, BR, NA |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z436190540
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, A1I3V, BR, NA |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: HEM, TES, FMT, NA |
 |
Olep In Complex With Lithocholic Acid In High Salt Crystallization Conditions
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-03 Classification: OXIDOREDUCTASE Ligands: HEM, 4OA, FMT, NA |
 |
Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With 1-(3,4-Dichlorobenzyl)-4-(((5-((4-Fluorophenethyl)Carbamoyl)Furan-2-Yl)Methyl)(4-Fluorophenyl)Carbamoyl)-1-(3-Phenylpropyl)Piperazin-1-Ium
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-04-03 Classification: OXIDOREDUCTASE Ligands: FAD, YJ6, PEG, IMD |
 |
Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With 4-(((5-((4-Fluorophenethyl)Carbamoyl)Furan-2-Yl)Methyl)(4-Fluorophenyl)Carbamoyl)-1-Methyl-1-(3-Phenylpropyl)Piperazin-1-Ium
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2024-04-03 Classification: OXIDOREDUCTASE Ligands: FAD, YIA, PEG, IMD, DMS |
 |
Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With 1-(3,4-Dichlorobenzyl)-4-(((5-((4-Fluorophenethyl)Carbamoyl)Furan-2-Yl)Methyl)Carbamoyl)-1-(3-Phenylpropyl)Piperazin-1-Ium
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-04-03 Classification: OXIDOREDUCTASE Ligands: FAD, YJK, PEG, DMS |
 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Half-Closed Conformation
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
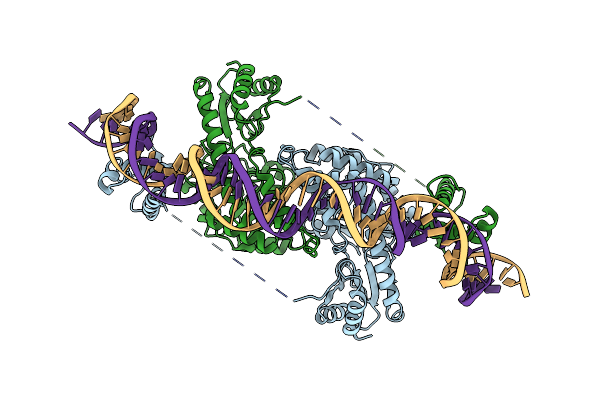 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Closed Conformation, C2 Symmetry.
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
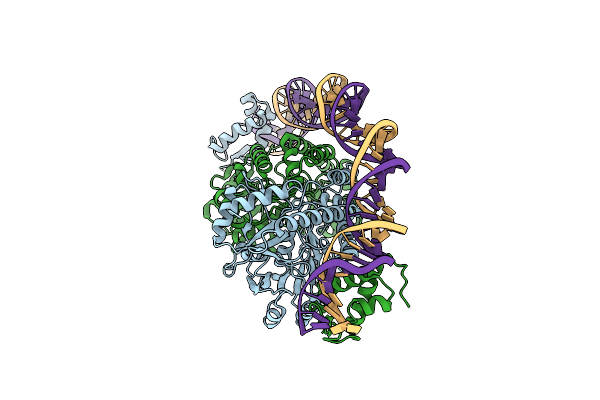 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Closed Conformation, C1 Symmetry
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
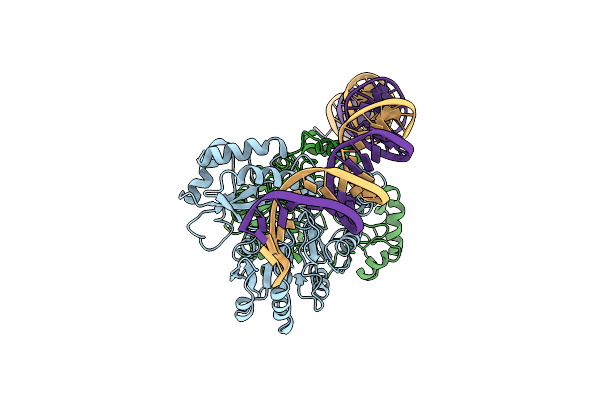 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Open Conformation
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-03-01 Classification: OXIDOREDUCTASE Ligands: FAD, DMS, MG, BR |
 |
Organism: Alkalihalobacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-09-28 Classification: DNA BINDING PROTEIN Ligands: CA, CL, PGE, EDO, PEG, P6G |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-07-13 Classification: OXYGEN TRANSPORT Ligands: HEM, SO4, GOL, IPA, BTB |

