Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Crystal Structure Of Repeats-In-Toxin-Like Domain From Aeromonas Hydrophila
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-14 Classification: CELL ADHESION Ligands: EDO, CA |
 |
Crystal Structure Of Vwfa Domain From Large Adhesion Protein Of Aeromonas Hydrophila
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-05-14 Classification: CELL ADHESION Ligands: CA, GOL |
 |
Fucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: FUL, CA, EDO, FUC |
 |
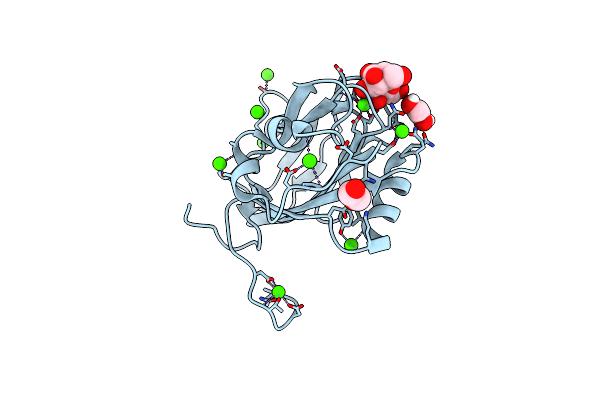
Allose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, VDS, VDV |
 |
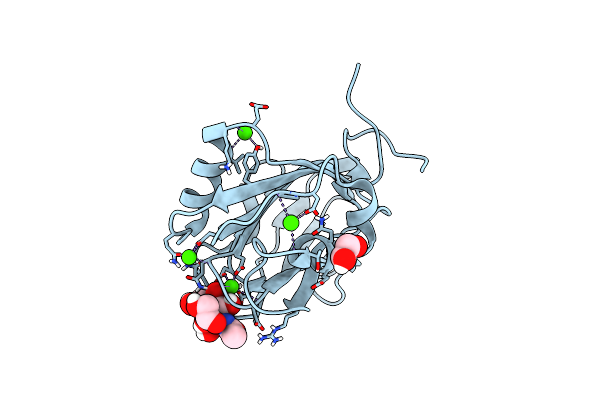
Mannose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, MAN, BMA |
 |
N-Acetyl-Glucosamine-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: NDG, NAG, CA, EDO |
 |
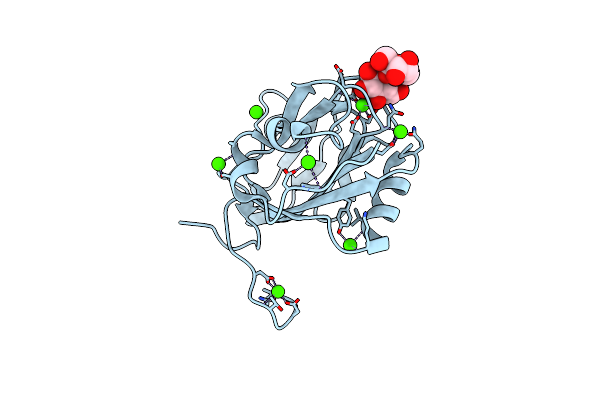
Inositol-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, INS |
 |
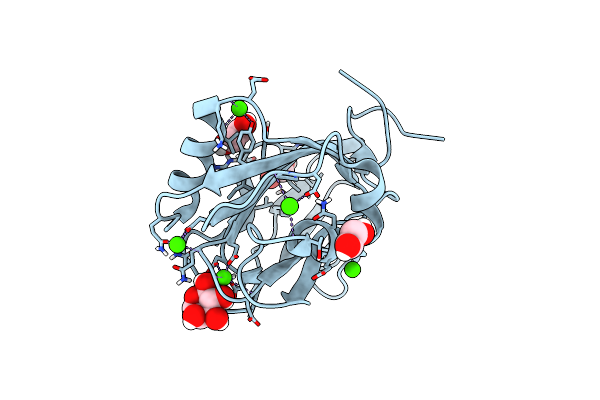
Sucrose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Arabinose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, ARA, EDO |
 |
Ribose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: RIP, CA, EDO |
 |
2-Deoxy-Glucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: Z61, CA, EDO |
 |
3-O-Methyl-Glucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, EDO, 3MG |
 |
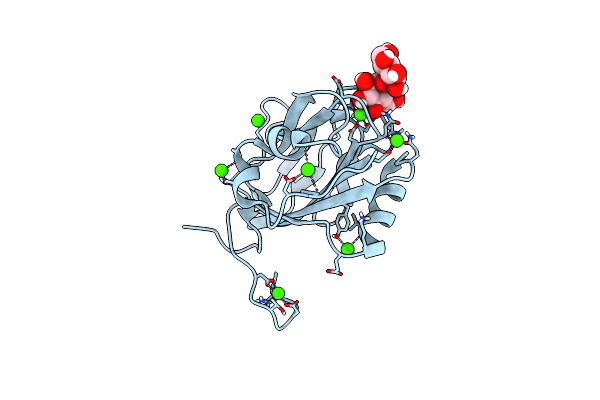
2-Deoxyribose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: UZJ, CA, EDO |
 |
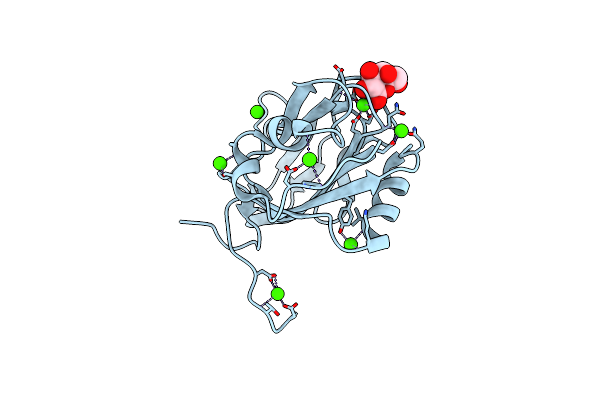
Trehalose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
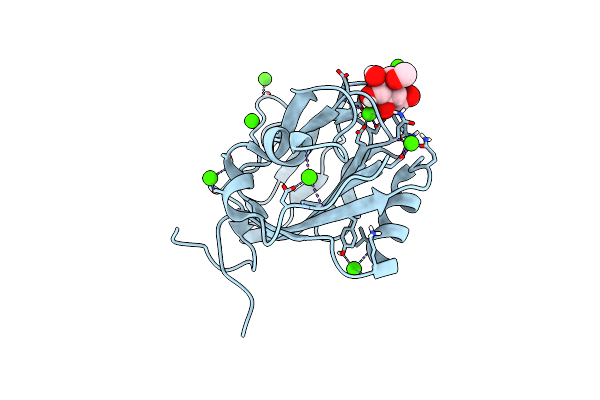
Galactose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: GAL, CA |
 |
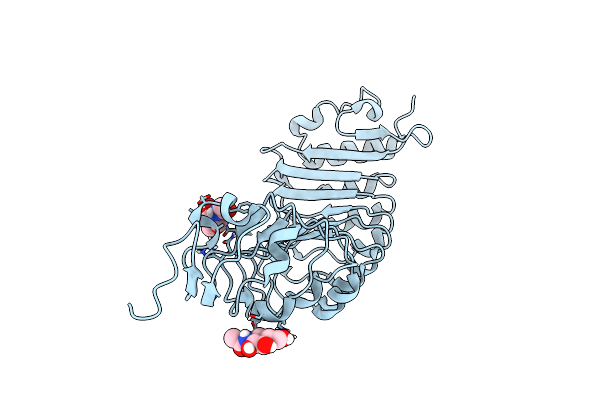
Alpha-Methyl-Glucose-Bound Structure Of Marinomonas Primoryensis Pa14 Carbohydrate-Binding Domain
Organism: Marinomonas primoryensis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-06-02 Classification: SUGAR BINDING PROTEIN Ligands: CA, GYP |
 |
Organism: Daucus carota
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2021-01-20 Classification: ANTIFREEZE PROTEIN Ligands: NAG |
 |
Organism: Rhagium mordax
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-08-26 Classification: ANTIFREEZE PROTEIN Ligands: EDO |
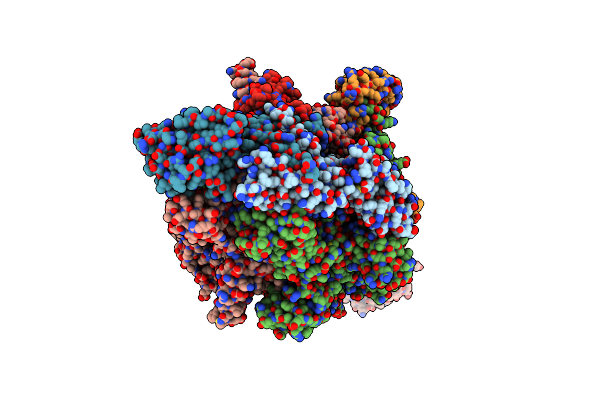 |
Thermus Thermophiles Rna Polymerase In Complex With Promoter Dna And Antibiotic Kanglemycin A
Organism: Thermus thermophilus (strain hb27 / atcc baa-163 / dsm 7039), Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2018-07-25 Classification: transcription/dna/antibiotic Ligands: KNG, ZN, MG |
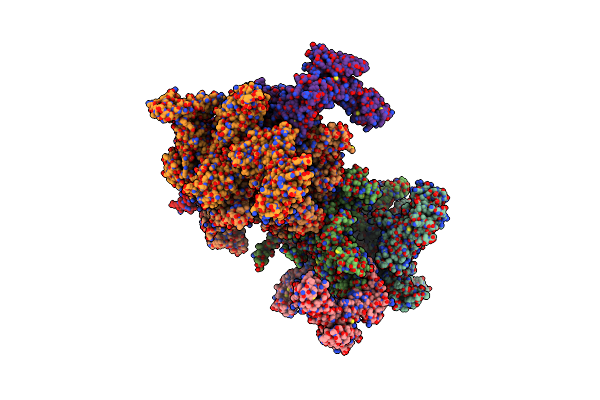 |
Escherichia Coli Rpob S531L Mutant Rna Polymerase Holoenzyme In Complex With Kanglemycin A
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:4.10 Å Release Date: 2018-07-25 Classification: transcription/antibiotic Ligands: KNG, MG, ZN |