Search Count: 254
 |
Organism: Clostridium carboxidivorans p7
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2024-03-13 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Gentamicin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: LLL, EDO, MG, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Tobramycin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: TOY, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Kanamycin B And Coenzyme A
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: 9CS, COA, EDO, SO4, CL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Paromomycin And Coenzyme A
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: COA, PAR, GOL |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma, Complex With Neomycin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: NMY, EDO |
 |
Crystal Structure Of Aminoglycoside Resistance Enzyme Apma H135A Mutant, Complex With Tobramycin And Coenzyme A
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: TOY, EDO, COA |
 |
Organism: Enterococcus faecalis og1rf
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2022-10-12 Classification: ANTIFUNGAL PROTEIN |
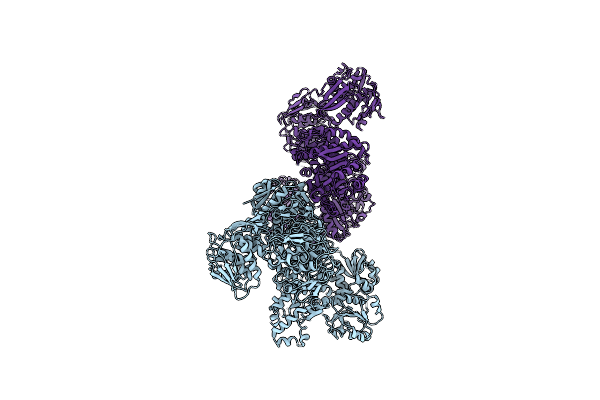 |
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: BIOSYNTHETIC PROTEIN |
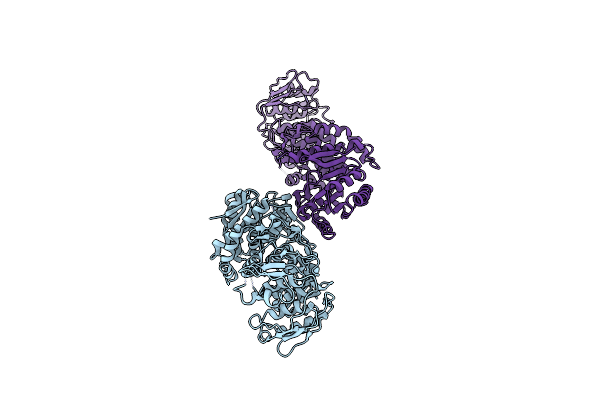 |
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: BIOSYNTHETIC PROTEIN |
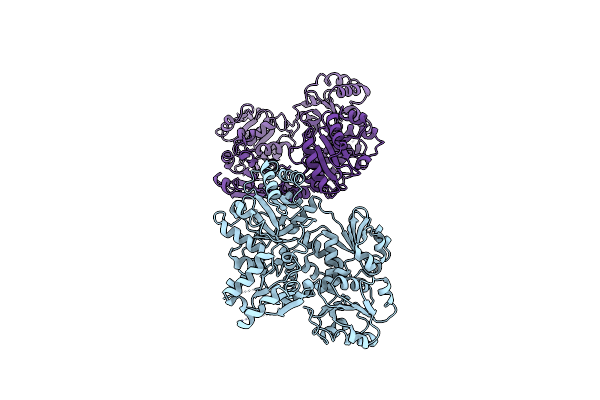 |
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2022-05-04 Classification: BIOSYNTHETIC PROTEIN |
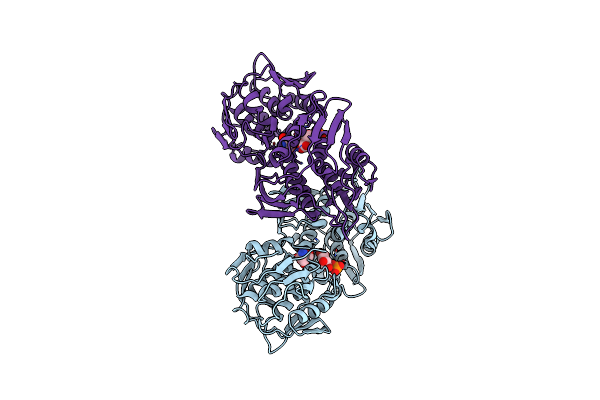 |
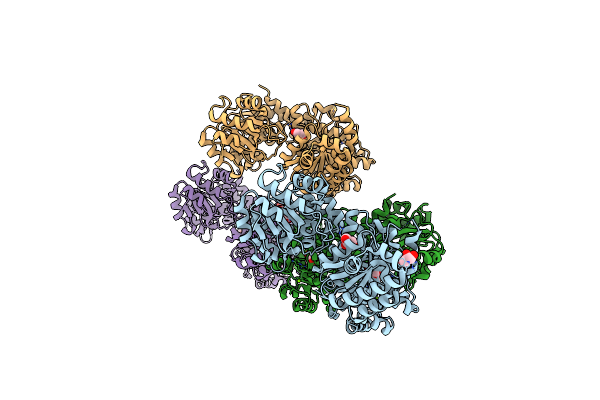
Crystal Structure Of The 5-Enolpyruvate-Shikimate-3-Phosphate Synthase (Epsps) Domain Of Aro1 From Candida Albicans In Complex With Shikimate-3-Phosphate
Organism: Candida albicans ca6
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-03-16 Classification: TRANSFERASE Ligands: TRS, S3P |
 |
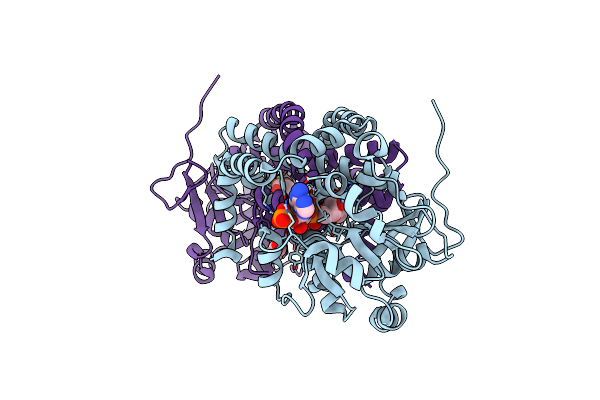
Crystal Structure Of The Shikimate Kinase + 3-Dehydroquinate Dehydratase + 3-Dehydroshikimate Dehydrogenase Domains Of Aro1 From Candida Albicans
Organism: Candida albicans ca6
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-16 Classification: TRANSFERASE,OXIDOREDUCTASE Ligands: MG, CL, GOL |
 |
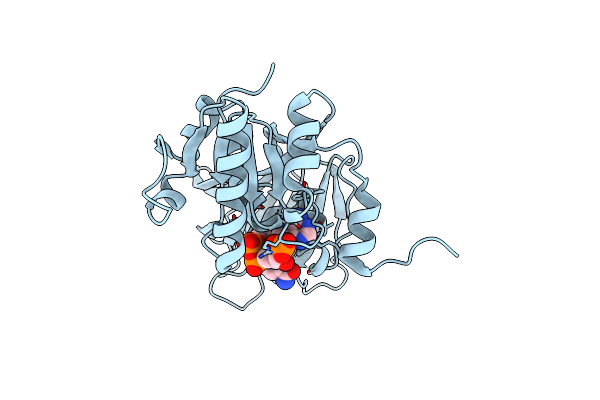
Crystal Structure Of The Mitochondrial Ketol-Acid Reductoisomerase Ilvc From Candida Auris
Organism: [candida] auris
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2022-02-16 Classification: ISOMERASE Ligands: NDP, MLI, ACY, MG |
 |
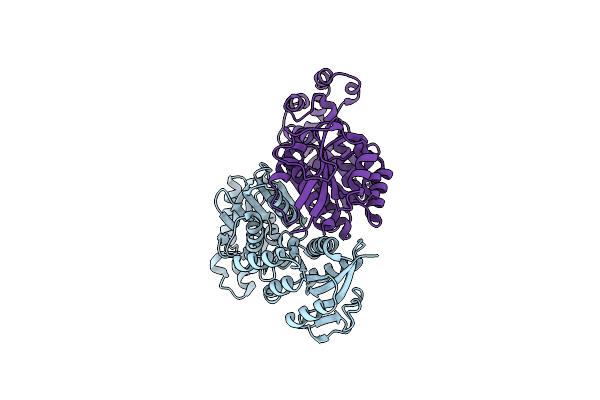
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2021-04-21 Classification: TRANSFERASE Ligands: NAP |
 |
Crystal Structure Of Unknown Function Protein Protein B9J08_000055 Candida Auris
Organism: Candida auris
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2021-04-07 Classification: STRUCTURAL GENOMICS |
 |
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2021-02-03 Classification: TRANSFERASE Ligands: TAR, FMT, CL |
 |
Organism: Serratia marcescens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2021-01-20 Classification: TRANSFERASE Ligands: EPE, MG |
 |
Crystal Structure Of Meta-Aac0038, An Environmental Aminoglycoside Resistance Enzyme, Mutant H168A In Complex With Apramycin And Coa
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2020-10-21 Classification: TRANSFERASE/ANTIBIOTIC Ligands: GOL, COA, AM2, CL |
 |
Crystal Structure Of Glycyl Radical Enzyme Ecl_02896 From Enterobacter Cloacae Subsp. Cloacae
Organism: Enterobacter cloacae subsp. cloacae (strain atcc 13047 / dsm 30054 / nbrc 13535 / ncdc 279-56)
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2020-08-12 Classification: LYASE Ligands: PEG, EDO |

