Search Count: 20
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And 5-Methoxy-1H-Indole-2-Carboxylic Acid
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: CTP, CA, VEO |
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And 2-(5-Bromo-1-(4-Hydroxyphenyl)-1H-Indol-3-Yl)-2-Oxoacetic Acid
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: CTP, CA, VE6 |
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And 2-(1-(4-Hydroxyphenyl)-5-Phenyl-1H-Indol-3-Yl)-2-Oxoacetic Acid
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: CTP, CA, W52, ACT |
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And 2-(1-(3,4-Dihydroxyphenyl)-5-Phenyl-1H-Indol-3-Yl)-2-Oxoacetic Acid
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: CTP, CA, W4X |
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And 2-(1-(3,4-Dihydroxyphenyl)-5-(3-Hydroxyphenyl)-1H-Indol-3-Yl)-2-Oxoacetic Acid
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2024-05-08 Classification: LIGASE Ligands: CTP, CA, W5E, ACT |
 |
Organism: Elizabethkingia anophelis ag1
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-02-07 Classification: TOXIN Ligands: SO4, GOL, EDO, PG4, CA, NA |
 |
Cryo-Em Structure Of The E.Coli 70S Ribosome In Complex With The Antibiotic Myxovalargin B.
Organism: Myxococcus fulvus, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: RIBOSOME Ligands: MG, ZN, FME, SPD |
 |
Cryo-Em Structure Of The E.Coli 50S Ribosomal Subunit In Complex With The Antibiotic Myxovalargin A.
Organism: Myxococcus fulvus, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Elizabethkingia anophelis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-04-21 Classification: TOXIN |
 |
Crystal Structure Of Mycobacterium Smegmatis Coabc In Complex With Ctp And Fmn
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-11-25 Classification: LIGASE Ligands: CTP, EDO, MG, FMN |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2020-11-25 Classification: LIGASE Ligands: CTP, CA, ACT, MES |
 |
Crystal Structure Of Mycobacterium Smegmatis Coab In Complex With Ctp And (4-Hydroxyphenyl)(2,3,4-Trihydroxyphenyl)Methanone
Organism: Mycolicibacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2020-11-25 Classification: LIGASE Ligands: CTP, CA, N9N, ACT |
 |
Cobalamin-Dependent Methionine Synthase (Meth) From Thermotoga Maritima (Oxidized, Monoclinic)
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-03-23 Classification: TRANSFERASE |
 |
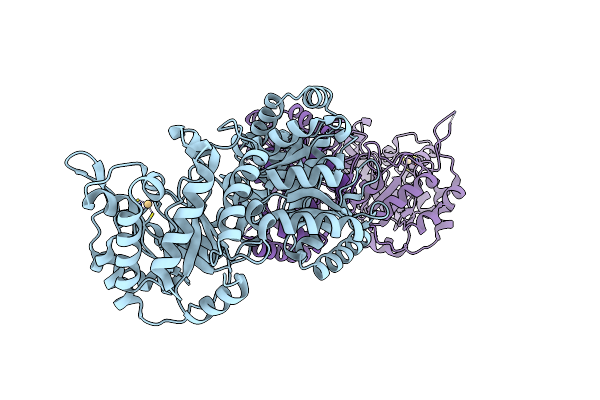
Cobalamin-Dependent Methionine Synthase (1-566) From T. Maritima (Oxidized, Orthorhombic)
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2004-03-23 Classification: TRANSFERASE |
 |
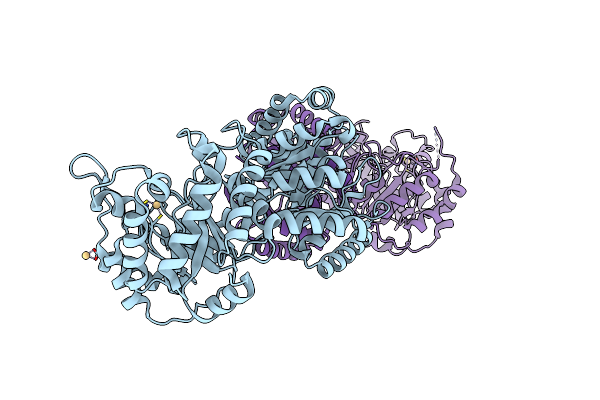
Cobalamin-Dependent Methionine Synthase (1-566) From Thermotoga Maritima (Cd2+ Complex)
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-03-23 Classification: TRANSFERASE Ligands: CD |
 |
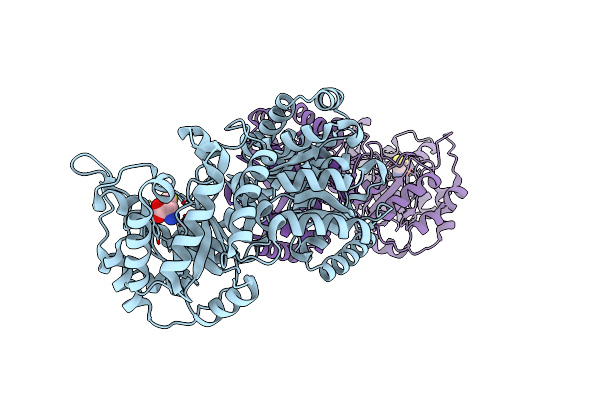
Cobalamin-Dependent Methionine Synthase (1-566) From Thermotoga Maritima (Cd2+ Complex, Se-Met)
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-03-23 Classification: TRANSFERASE Ligands: CD |
 |
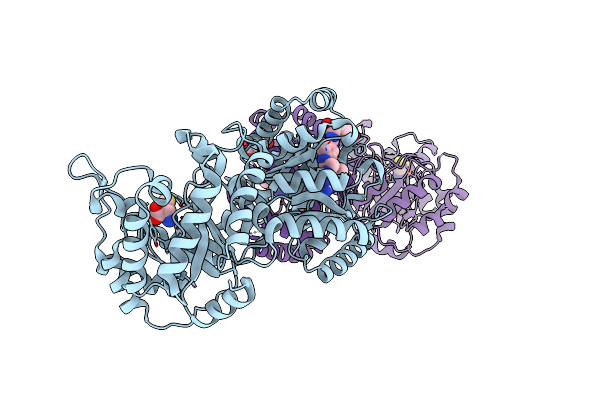
Cobalamin-Dependent Methionine Synthase (1-566) From Thermotoga Maritima (Cd2+:L-Hcy Complex, Se-Met)
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-03-23 Classification: TRANSFERASE Ligands: CD, HCS |
 |
Cobalamin-Dependent Methionine Synthase (1-566) From Thermotoga Maritima (Cd2+, Hcy, Methyltetrahydrofolate Complex)
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-03-23 Classification: TRANSFERASE Ligands: CD, HCS, C2F |
 |
Oxidized Homo Sapiens Betaine-Homocysteine S-Methyltransferase In Complex With Four Sm(Iii) Ions
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2002-09-11 Classification: TRANSFERASE Ligands: SM, CIT |
 |
Reduced Homo Sapiens Betaine-Homocysteine S-Methyltransferase In Complex With S-(Delta-Carboxybutyl)-L-Homocysteine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2002-09-11 Classification: TRANSFERASE Ligands: ZN, CBH, CIT |

