Search Count: 10
 |
Organism: Colysis wrightii
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-11-08 Classification: PLANT PROTEIN Ligands: GOL |
 |
Organism: Pseudomonas monteilii
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-05-31 Classification: TOXIN Ligands: MG |
 |
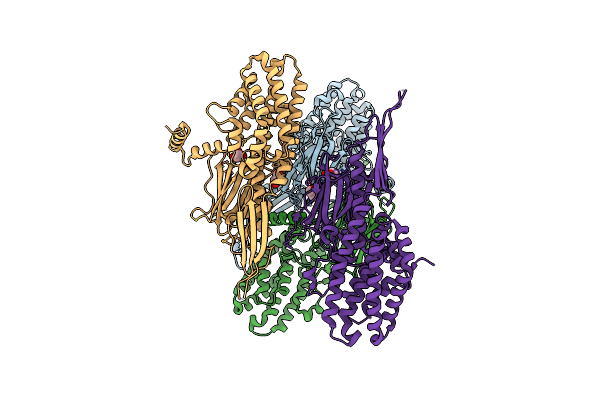
Organism: Pseudomonas monteilii
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: TOXIN Ligands: MG |
 |
Organism: Pseudomonas monteilii
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: TOXIN Ligands: MG |
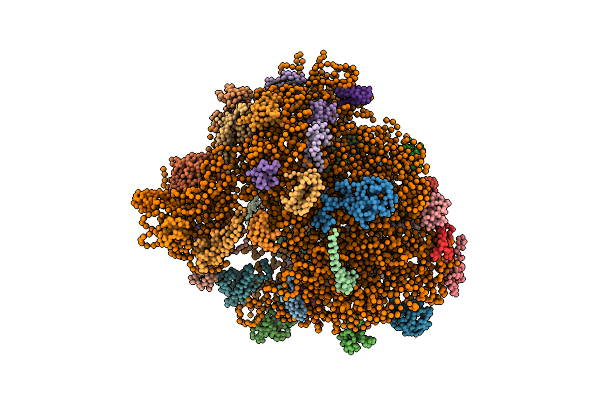 |
Real Space Refined Coordinates Of The 30S And 50S Subunits Fitted Into The Low Resolution Cryo-Em Map Of The Ef-G.Gtp State Of E. Coli 70S Ribosome
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |
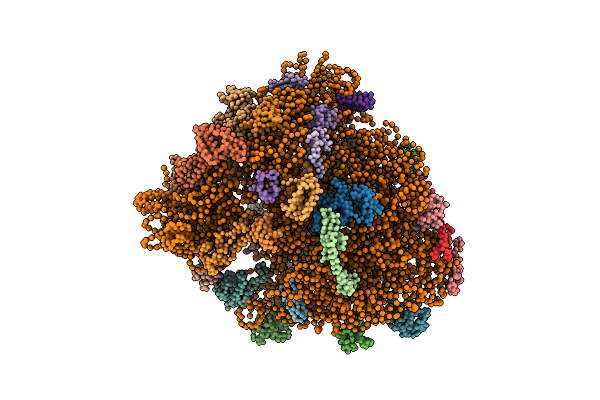 |
Real Space Refined Coordinates Of The 30S And 50S Subunits Fitted Into The Low Resolution Cryo-Em Map Of The Initiation-Like State Of E. Coli 70S Ribosome
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |
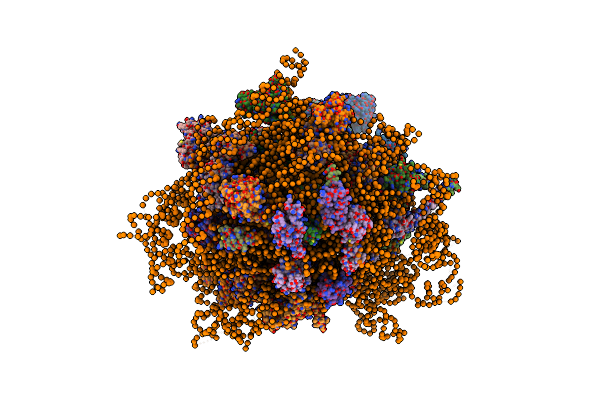 |
Structure Of A Mammalian 80S Ribosome Obtained By Docking Homology Models Of The Rna And Proteins Into An 8.7 A Cryo-Em Map
Organism: Canis familiaris, Canis familiaris
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOMAL PROTEIN/RNA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-08-22 Classification: OXIDOREDUCTASE, LYASE Ligands: FE2 |
 |
Structural Studies Of Cholesterol Biosynthesis: Mevalonate 5-Diphosphate Decarboxylase And Isopentenyl Diphosphate Isomerase
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2001-03-28 Classification: ISOMERASE Ligands: MN |
 |
The X-Ray Crystal Structure Of Mevalonate 5-Diphosphate Decarboxylase At 2.3 Angstrom Resolution.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2001-03-21 Classification: LYASE |

