Search Count: 33
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2020-09-16 Classification: METAL BINDING PROTEIN Ligands: HEC, MPD, SO4 |
 |
Fe-Bound Structure Of An Engineered Metal-Dependent Protein Trimer, Tricyt2
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-16 Classification: METAL BINDING PROTEIN Ligands: HEC, FE2, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-16 Classification: METAL BINDING PROTEIN Ligands: HEC, MN, CL, CA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-16 Classification: METAL BINDING PROTEIN Ligands: HEC, CO, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-09-16 Classification: METAL BINDING PROTEIN Ligands: HEC, CL, CU, PEG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-09-16 Classification: METAL BINDING PROTEIN Ligands: HEC, MN, CL, CA |
 |
Ni-Bound Structure Of An Engineered Metal-Dependent Protein Trimer, Tricyt1
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-09-16 Classification: METAL BINDING PROTEIN Ligands: HEC, NI, NA, CL, MPD |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-09-16 Classification: METAL BINDING PROTEIN Ligands: HEC, NI, CA, CL |
 |
Co-Bound Structure Of An Engineered Protein Trimer, Tricyt3, With Delta Isomerism At The Hexahistidine Coordination Site
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-16 Classification: METAL BINDING PROTEIN Ligands: HEC, CO, CL, MG |
 |
Cu-Bound Structure Of An Engineered Metal-Dependent Protein Trimer, Tricyt1
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2020-09-16 Classification: METAL BINDING PROTEIN Ligands: HEC, CU, CA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-01-29 Classification: METAL BINDING PROTEIN Ligands: HEC, HAE, FE, ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-01-29 Classification: METAL BINDING PROTEIN Ligands: HEC, ZN, FE, HAE, 1PE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-01-29 Classification: METAL BINDING PROTEIN Ligands: HEC, ZN, FE, HAE |
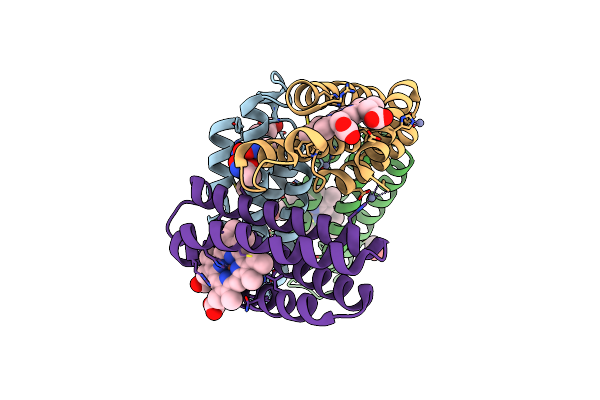 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-01-29 Classification: METAL BINDING PROTEIN Ligands: ZN, HAE, HEC, FE |
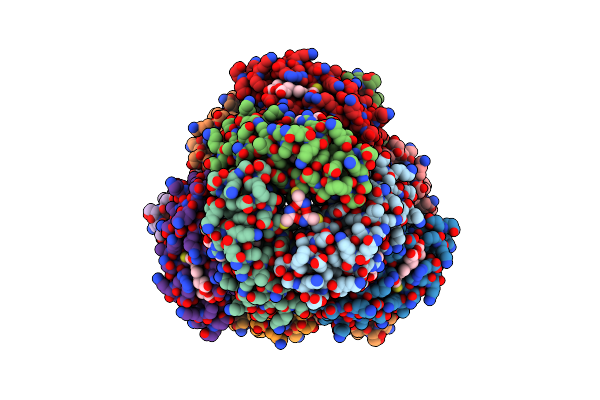 |
Cryo-Em Structure Of Bimetallic Dodecameric Cage Design 3 (Bmc3) From Cytochrome Cb562
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2020-01-29 Classification: METAL BINDING PROTEIN Ligands: HEC, HAE, ZN, FE |
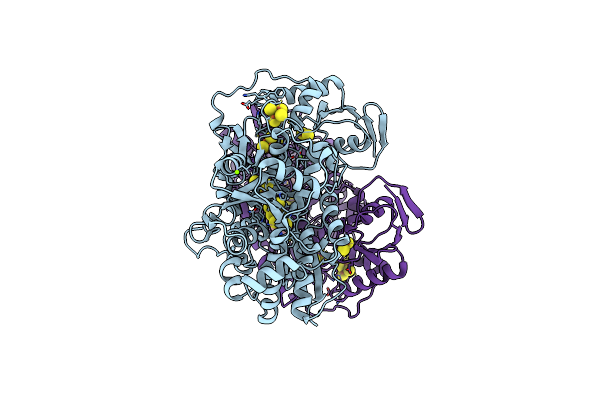 |
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, MG |
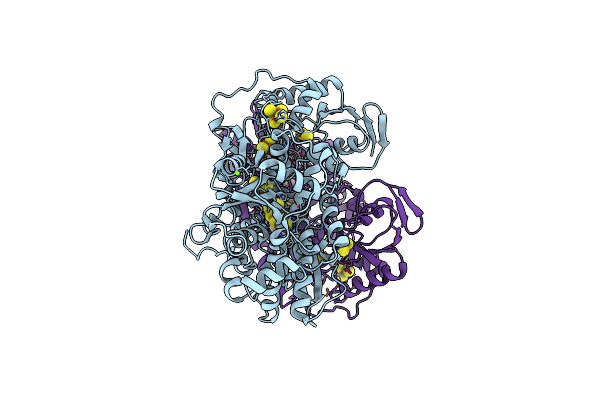 |
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, MG |
 |
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, MG |
 |
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-11-07 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, MG |
 |
Organism: Clostridium pasteurianum
Method: X-RAY DIFFRACTION Release Date: 2018-11-07 Classification: OXIDOREDUCTASE Ligands: 402, SF4, FES, MG |

