Search Count: 15
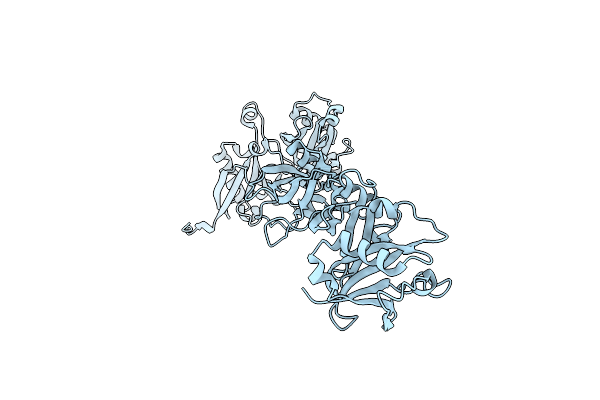 |
Mechanism Of Usp7 (Hausp) Activation By Its C-Terminal Ubiquitin-Like Domain (Hubl) And Allosteric Regulation By Gmp-Synthetase.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-10-19 Classification: HYDROLASE Ligands: CL |
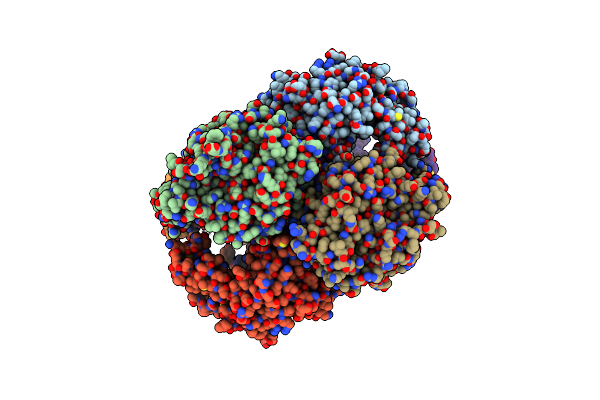 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-04-06 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-02-12 Classification: HYDROLASE Ligands: ZN, SO4, MES, GOL |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2008-02-12 Classification: HYDROLASE Ligands: G2F, ZN, SO4, MES, GOL |
 |
Organism: Sorghum bicolor
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-05-20 Classification: HYDROLASE |
 |
Organism: Sorghum bicolor milo
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-05-20 Classification: HYDROLASE Ligands: BGC, CCN, IPH |
 |
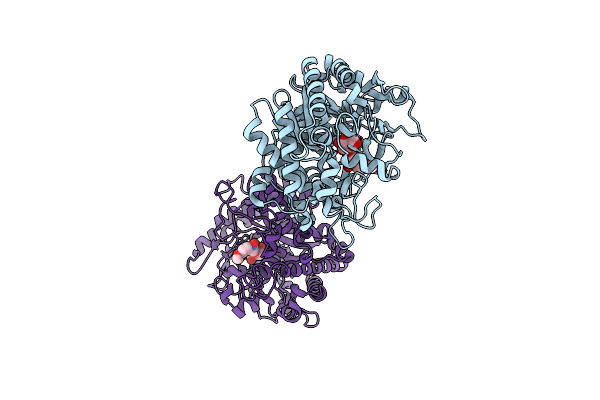
Crystal Structure Of The Zea Maze Beta-Glucosidase-1 In Complex With Gluco-Tetrazole
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-05-20 Classification: HYDROLASE Ligands: NTZ |
 |
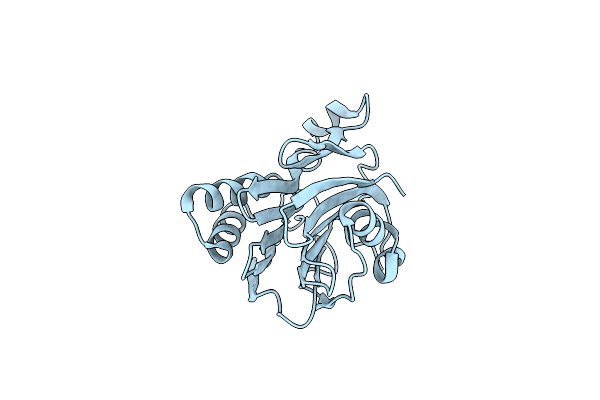
Crystal Structure Of The Inactive Double Mutant Of The Maize Beta-Glucosidase Zmglu1-E191D-F198V In Complex With Dimboa-Glucoside
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2003-03-11 Classification: HYDROLASE Ligands: HBO, BGC |
 |
Crystal Structure Of Methanobacterium Thermoautotrophicum Conserved Protein Mth1020 Reveals An Ntn-Hydrolase Fold
Organism: Methanothermobacter
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-05-29 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2001-02-19 Classification: HYDROLASE |
 |
Crystal Structure Of A Monocot (Maize Zmglu1) Beta-Glucosidase In Complex With P-Nitrophenyl-Beta-D-Thioglucoside
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2001-02-19 Classification: HYDROLASE Ligands: PSG |
 |
Crystal Structure Of The Inactive Mutant Monocot (Maize Zmglu1) Beta-Glucosidase Zm Glu191Asp
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2000-12-11 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of The Inactive Mutant Monocot (Maize Zmglu1) Beta-Glucosidase Zmglue191D In Complex With The Natural Aglycone Dimboa
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-12-11 Classification: HYDROLASE Ligands: HBO |
 |
Crystal Structure Of The Inactive Mutant Monocot (Maize Zmglu1) Beta-Glucosidase Zmglue191D In Complex With The Competitive Inhibitor Dhurrin
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-12-11 Classification: HYDROLASE Ligands: BGC, DHR |
 |
Crystal Structure Of The Inactive Mutant Monocot (Maize Zmglu1) Beta-Glucosidase Zmglue191D In Complex With The Natural Substrate Dimboa-Beta-D-Glucoside
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2000-12-11 Classification: HYDROLASE Ligands: BGC, HBO |

