Search Count: 95
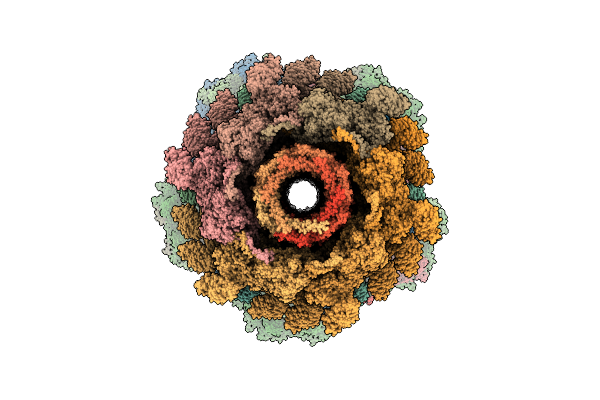 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRAL PROTEIN |
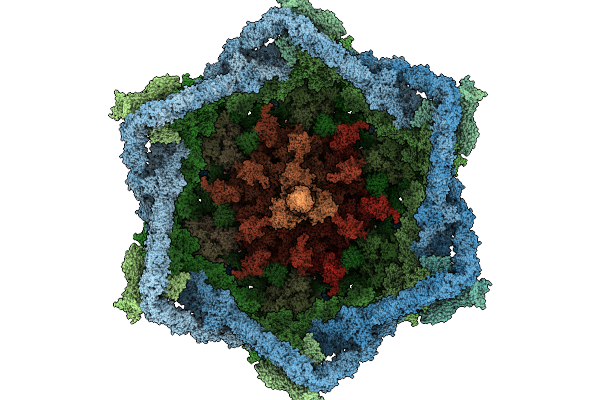 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRAL PROTEIN Ligands: ZN |
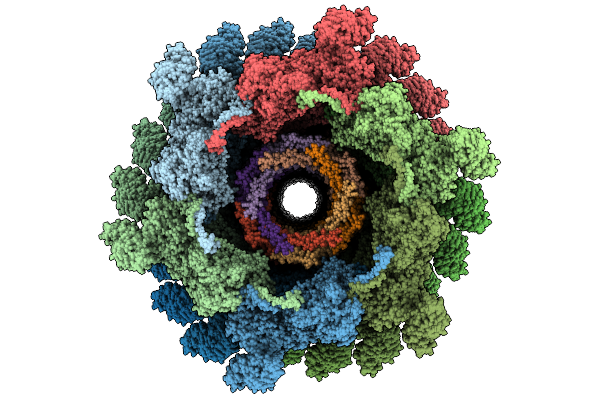 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRAL PROTEIN |
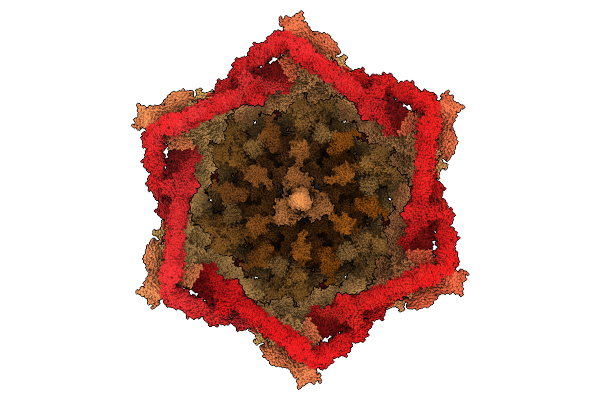 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Spitrobot-2 Advances Time-Resolvedcryo-Trapping Crystallography To Under 25 Ms: T4 Lysozyme, Mutant L99A (Apo State)
Organism: Escherichia phage t4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-12-03 Classification: HYDROLASE |
 |
Spitrobot-2 Advances Time-Resolvedcryo-Trapping Crystallography To Under 25 Ms: T4 Lysozyme, Mutant L99A Bound With Indole (1 S Soaking)
Organism: Escherichia phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-12-03 Classification: HYDROLASE Ligands: IND |
 |
Spitrobot-2 Advances Time-Resolvedcryo-Trapping Crystallography To Under 25 Ms: T4 Lysozyme, Mutant L99A Bound With Indole (10 S Soaking)
Organism: Escherichia phage t4
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-12-03 Classification: HYDROLASE Ligands: IND, NA |
 |
T4 Bacteriophage Replicative Polymerase Captured In Polymerase Exchange State 1
Organism: Escherichia phage t4, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: replication/DNA |
 |
T4 Bacteriophage Replicative Polymerase Captured In Polymerase Exchange State 2
Organism: Escherichia phage t4, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: replication/DNA |
 |
T4 Bacteriophage Replicative Holoenzyme Containing Triple Mutations D75R, Q430E, And K432E In The Exonuclease-Deficient Polymerase
Organism: Escherichia phage t4, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: Transferase/DNA |
 |
T4 Bacteriophage Replicative Polymerase Captured In Polymerase Exchange State 3
Organism: Escherichia phage t4, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: Replication/DNA Ligands: CA, D3T |
 |
Structure Of The Bacterial Ribosome Without Hypoxia-Induced Rrna Modifications
Organism: Escherichia phage t4, Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG |
 |
Structure Of The Bacterial Ribosome With Hypoxia-Induced Rrna Modifications
Organism: Escherichia phage t4, Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG |
 |
Cryo-Em Structure Of Linear Intron Of Thymidylate Synthase (Td) Gene Of Bacteriophage T4
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA Ligands: MG |
 |
Cryo-Em Structure Of Circular Intron Of Thymidylate Synthase (Td) Gene Of Bacteriophage T4
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA Ligands: MG |
 |
Organism: Escherichia phage t4, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: ISOMERASE Ligands: MG |
 |
Organism: Escherichia phage t4, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: ISOMERASE/DNA |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With O-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1F, ZN, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With C-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.50A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1J, ZN, SF4 |
 |
Crystal Structure Of The Wild-Type Thermus Thermophilus 70S Ribosome In Complex With Se-Cresomycin, Mrna, Deacylated A-Site Trnaphe, Aminoacylated P-Site Fmet-Trnamet, And Deacylated E-Site Trnaphe At 2.45A Resolution
Organism: Escherichia coli, Escherichia phage t4, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-08-27 Classification: RIBOSOME Ligands: MG, K, A1A1K, ZN, SF4 |

