Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
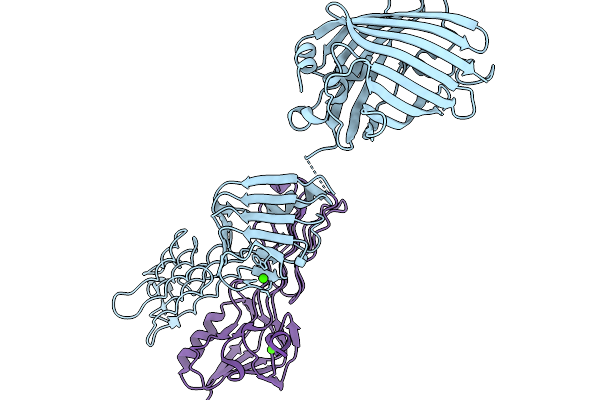 |
Organism: Aequorea victoria, Escherichia phage pp01
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: CELL ADHESION Ligands: CA |
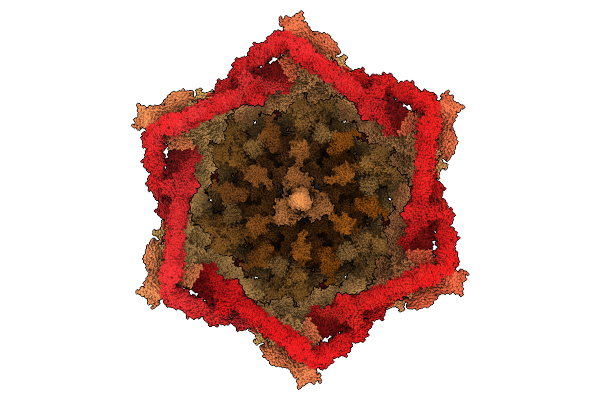 |
Organism: Escherichia phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Cryo-Em Structure Of Gp77 Within The In Vitro Reconstituted Razr:Gp77 Complex
Organism: Escherichia phage secphi27
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN |
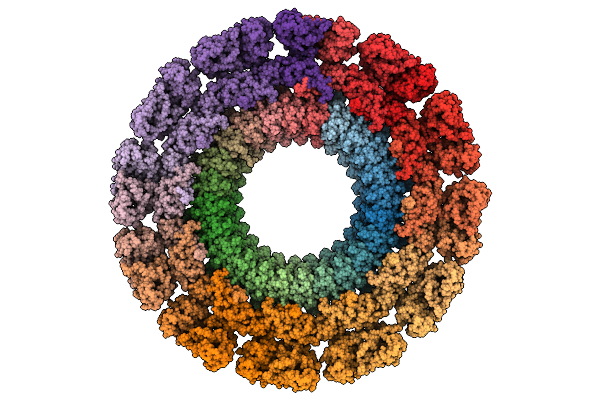 |
Cryo-Em Map Of The In Vitro Reconstituted Razr:Gp77 Complex With Alphafold-Predicted Models Fitted Into The Density.
Organism: Escherichia phage secphi27, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: ZN |
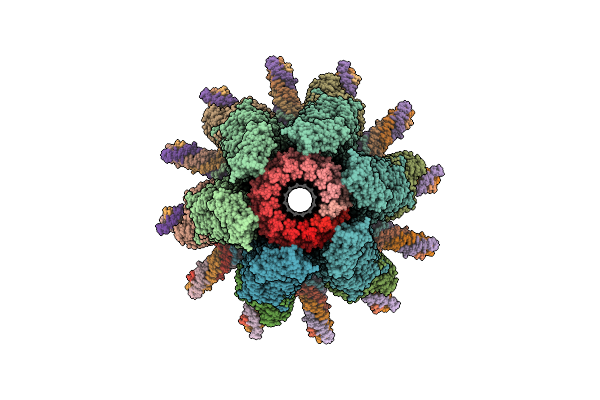 |
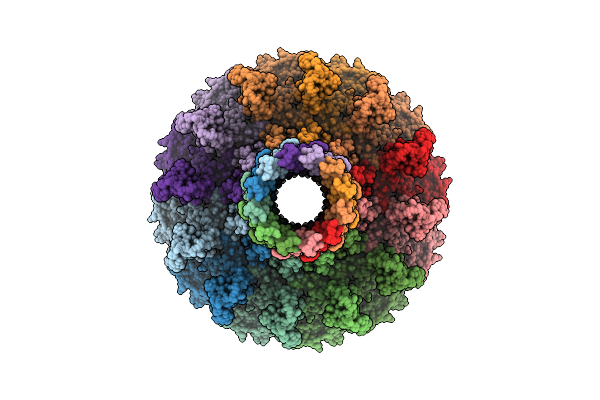
Organism: Escherichia phage n4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN |
 |
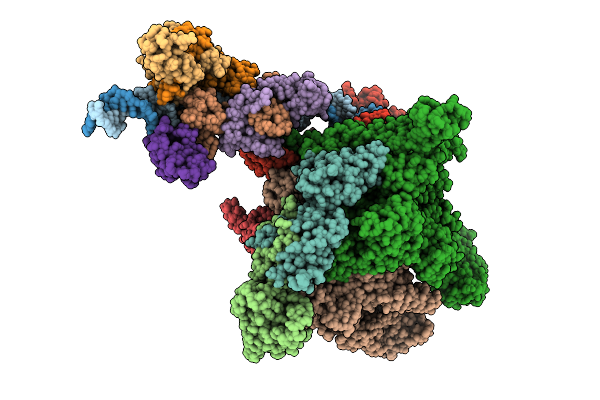
Organism: Escherichia phage n4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN |
 |
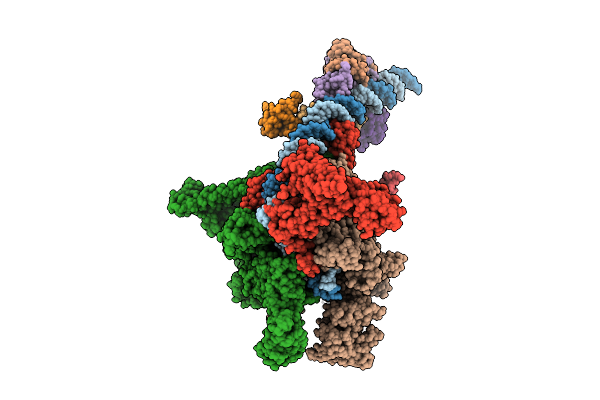
Cryo-Em Structure Of E.Coli Transcription Initiation Complex With Escherichia Phage Mu Late Transcription Activator C
Organism: Escherichia coli (strain k12), Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION/DNA |
 |
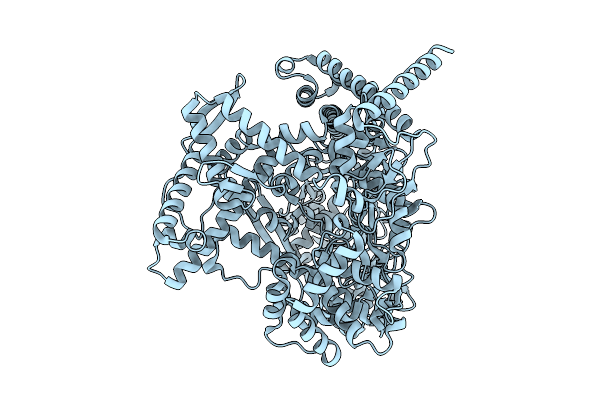
Cryo-Em Structure Of E.Coli Transcription Initiation Complex With Escherichia Phage Mu Middle Transcription Activator Mor
Organism: Escherichia coli (strain k12), Escherichia phage mu
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION/DNA |
 |
Organism: Escherichia phage n4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN, TRANSFERASE |
 |
Organism: Escherichia phage n4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN, TRANSFERASE |
 |
Organism: Escherichia phage n4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN, TRANSFERASE |
 |
Organism: Escherichia phage n4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN, TRANSFERASE |
 |
Organism: Escherichia phage n4
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN, TRANSFERASE |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde In Complex With Rtp45 Superinfection Exclusion Protein From Rtp Bacteriophage And Endogenous Lptm
Organism: Shigella flexneri, Escherichia phage rtp, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLM, PXS |
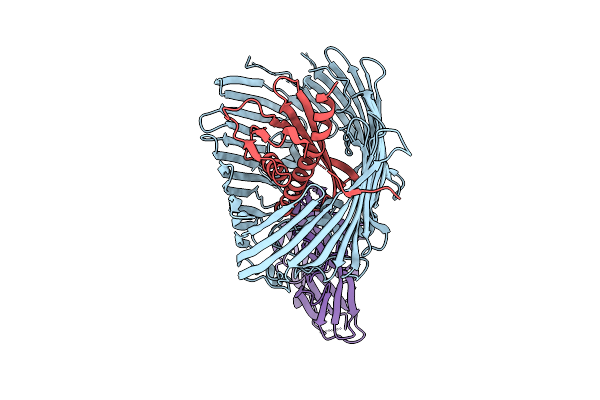 |
Cryo-Em Structure Of The Open State Of Shigella Flexneri Lptde Bound By The Rbp Of Oekolampad Phage
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
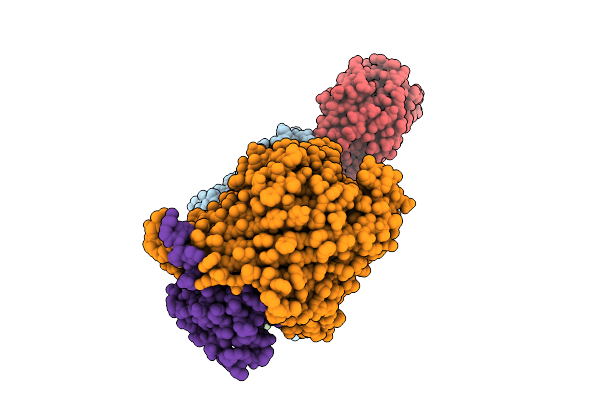 |
Cryo-Em Structure Of Shigella Flexneri Lptde Dimer: Closed-State Unbound And Open-State Bound By Oekolampad Phage Rbp
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
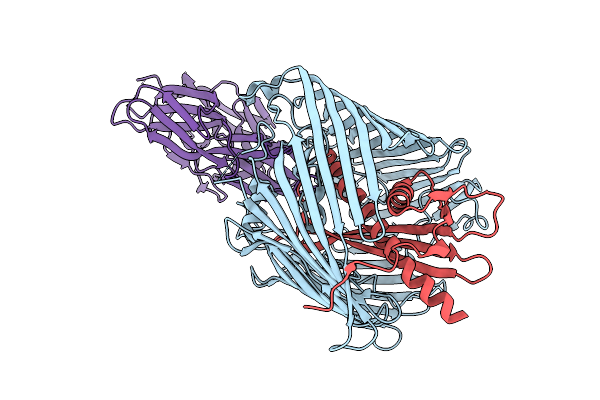 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By Phage Rbp Reveals N-Terminal Strand Insertion Into Lateral Gate
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
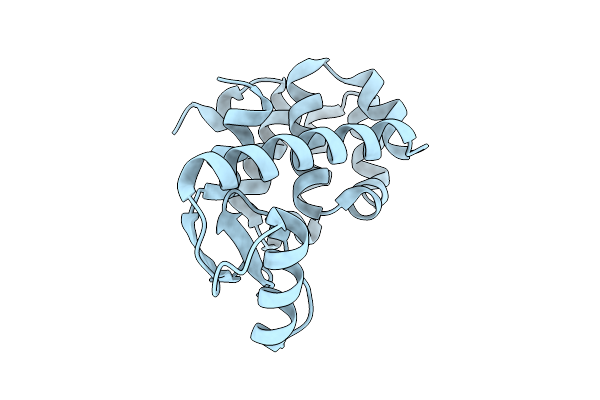 |
Spitrobot-2 Advances Time-Resolvedcryo-Trapping Crystallography To Under 25 Ms: T4 Lysozyme, Mutant L99A (Apo State)
Organism: Escherichia phage t4
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE |
 |
Spitrobot-2 Advances Time-Resolvedcryo-Trapping Crystallography To Under 25 Ms: T4 Lysozyme, Mutant L99A Bound With Indole (1 S Soaking)
Organism: Escherichia phage t4
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE Ligands: IND |
 |
Spitrobot-2 Advances Time-Resolvedcryo-Trapping Crystallography To Under 25 Ms: T4 Lysozyme, Mutant L99A Bound With Indole (10 S Soaking)
Organism: Escherichia phage t4
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: HYDROLASE Ligands: IND, NA |