Search Count: 21
 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: ELECTRON MICROSCOPY Release Date: 2022-09-21 Classification: PROTON TRANSPORT Ligands: SF4, FMN, FES, CA, 3PE, LFA |
 |
Complex I From E. Coli, Lmng-Purified, Under Turnover At Ph 6, Closed State
Organism: Escherichia coli str. k-12 substr. mc4100
Method: ELECTRON MICROSCOPY Release Date: 2022-09-21 Classification: PROTON TRANSPORT Ligands: SF4, FMN, NAI, FES, CA, 3PE, DCQ, LFA |
 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: ELECTRON MICROSCOPY Release Date: 2022-09-21 Classification: PROTON TRANSPORT Ligands: SF4, FMN, NAI, FES, CA, 3PE, LFA |
 |
Complex I From E. Coli, Lmng-Purified, Under Turnover At Ph 6, Open-Ready State
Organism: Escherichia coli str. k-12 substr. mc4100, Escherichia coliescherichia coli str. k-12 substr. mc4100
Method: ELECTRON MICROSCOPY Release Date: 2022-09-21 Classification: PROTON TRANSPORT Ligands: SF4, FMN, NAI, FES, CA, 3PE, LFA, C14 |
 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: FCO, NI, MG, SO4, SF4, F3S, SF3, CL |
 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: FCO, NI, MG, SO4, SF4, F3S, SF3, LMT, CL |
 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: NFV, MG, SF4, F3S, SF3, CL |
 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2015-11-25 Classification: OXIDOREDUCTASE Ligands: FCO, NI, MG, SO4, SF4, F3S, SF3, LMT, CL |
 |
Organism: Proteus vulgaris, Escherichia coli, Thermus thermophilus hb8, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2015-10-21 Classification: RIBOSOME Ligands: MG, ZN |
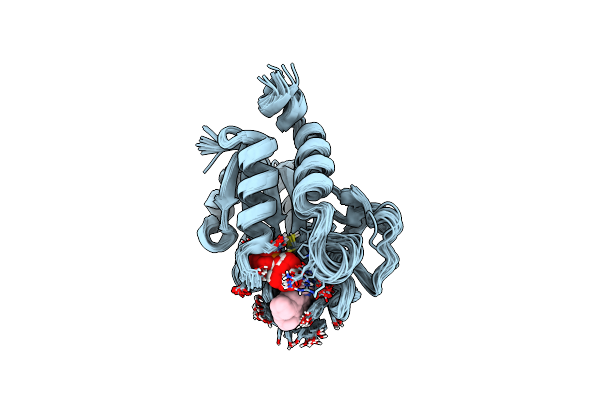 |
Organism: Escherichia coli str. k-12 substr. mc4100
Method: SOLUTION NMR Release Date: 2015-05-20 Classification: ELECTRON TRANSPORT Ligands: FMN |
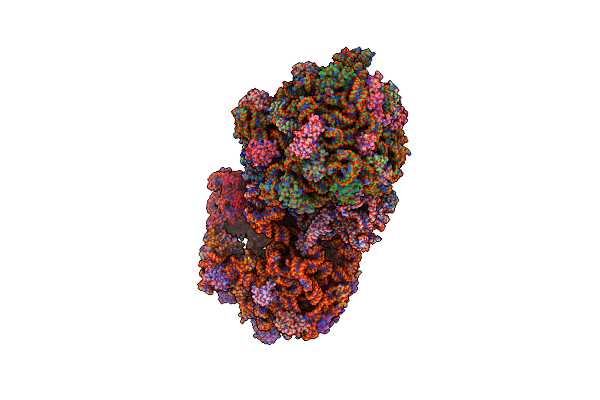 |
Organism: Synthetic construct, Thermus thermophilus hb8, Escherichia coli str. k-12 substr. mc4100, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2015-05-13 Classification: RIBOSOME Ligands: MG, PAR, ZN |
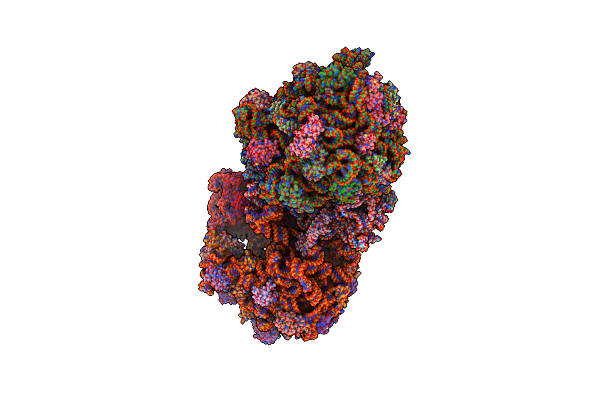 |
Organism: Synthetic construct, Thermus thermophilus hb8, Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2015-05-13 Classification: RIBOSOME Ligands: MG, ZN |
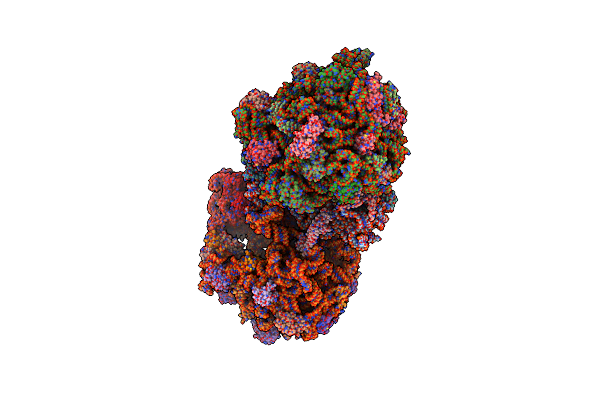 |
Organism: Synthetic construct, Thermus thermophilus hb8, Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2015-05-13 Classification: RIBOSOME Ligands: MG, PAR, ZN |
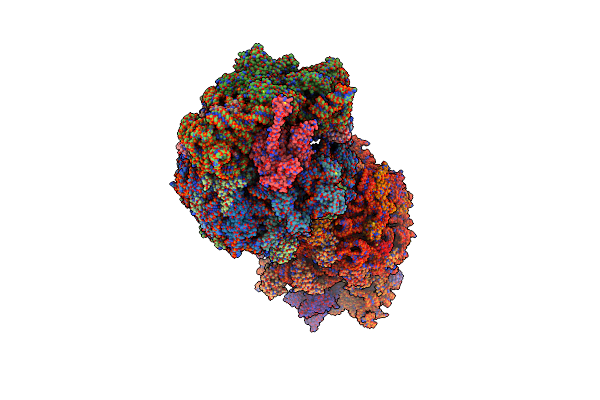 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G In The Pre-Translocational State
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: RIBOSOME Ligands: MG, K, ZN, SF4, GDP |
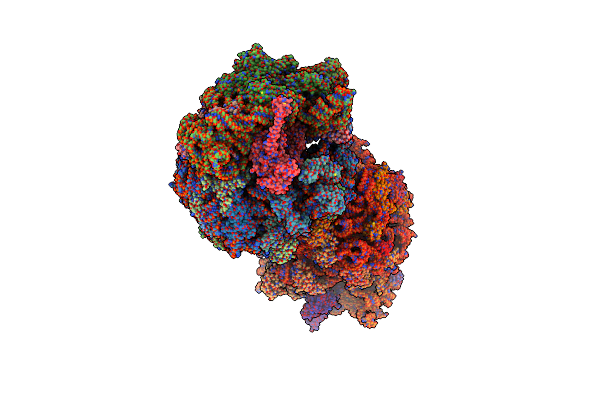 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G And Fusidic Acid In The Post-Translocational State
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: RIBOSOME Ligands: MG, ZN, SF4, FUA, GDP |
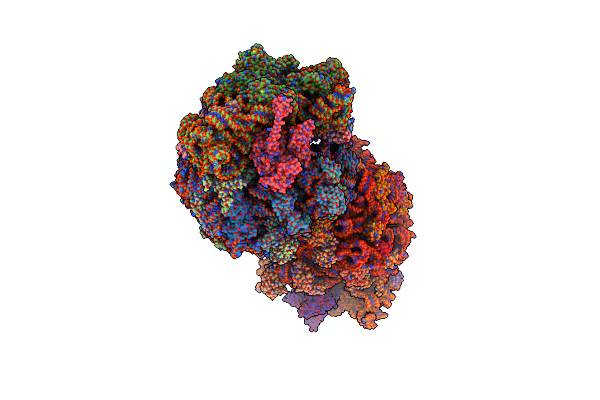 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G Trapped By The Antibiotic Dityromycin
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Streptomyces sp., Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: Ribosome/antibiotic Ligands: MG, ZN, SF4, GDP |
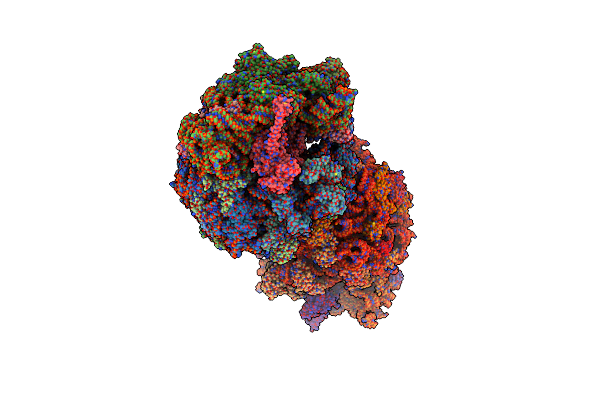 |
Crystal Structure Of The Thermus Thermophilus 70S Ribosome In Complex With Elongation Factor G In The Post-Translocational State (Without Fusitic Acid)
Organism: Escherichia coli str. k-12 substr. mc4100, Thermus thermophilus, Synthetic construct, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-01-28 Classification: RIBOSOME Ligands: MG, K, ZN, SF4, GDP |
 |
Structure Of The Curli Transport Lipoprotein Csgg In A Non-Lipidated, Pre-Pore Conformation
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-09-24 Classification: TRANSPORT PROTEIN |
 |
Structure Of The Curli Transport Lipoprotein Csgg In Its Membrane- Bound Conformation
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2014-09-24 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Escherichia Coli Protein Yoda In Complex With Ni - Artifact Of Purification.
Organism: Escherichia coli str. k-12 substr. mc4100
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-06-25 Classification: METAL BINDING PROTEIN Ligands: SO4, NI |

