Search Count: 22
 |
Escherichia Coli 70S Ribosome In Complex With Staphylococcus Aureus Fusb-Ef-G (Fusb-Ef-G-70S*)
Organism: Staphylococcus aureus, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:2.41 Å Release Date: 2025-03-26 Classification: RIBOSOME Ligands: ZN, K, MG |
 |
Organism: Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: IMMUNE SYSTEM Ligands: MG, NAD |
 |
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant F254I Complexed With Fe, Nad+, And Ethylene Glycol
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, FE |
 |
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant N151G, L259V Complexed With Fe, Nadh, And Glycerol (Absence Of Nicotinamide Ring)
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: APR, FE |
 |
Crystal Structure Of E.Coli Alcohol Dehydrogenase - Fuco Mutant F254I Complexed With Fe, Nadh, And Glycerol
Organism: Escherichia coli str. k-12 substr. mg1655
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-10-19 Classification: OXIDOREDUCTASE Ligands: NAI, APR, FE, GOL |
 |
Organism: Escherichia coli (strain k12), Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: RIBOSOME |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2021-01-13 Classification: LYASE |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2021-01-13 Classification: LYASE |
 |
Cryo-Em Structure Of Escherichia Coli Acrbz And Darpin In Saposin A-Nanodisc With Cardiolipin
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Escherichia Coli Acrbz And Darpin In Saposin A-Nanodisc
Organism: Escherichia coli k12, Synthetic construct, Shigella boydii 965-58
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Escherichia Coli Acrb And Darpin In Saposin A-Nanodisc With Cardiolipin
Organism: Escherichia coli k12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Escherichia Coli Acrb And Darpin In Saposin A-Nanodisc
Organism: Escherichia coli k12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli str. k-12 substr. mg1655star
Method: ELECTRON MICROSCOPY Release Date: 2020-01-22 Classification: SIGNALING PROTEIN |
 |
Structure Of The Antibacterial Peptide Abc Transporter Mcjd In An Apo Inward Occluded Conformation
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:4.71 Å Release Date: 2017-09-06 Classification: MEMBRANE PROTEIN |
 |
Structure Of The Antibacterial Peptide Abc Transporter Mcjd In A High Energy Outward Occluded Intermediate State
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2017-09-06 Classification: MEMBRANE PROTEIN Ligands: ADP, MG, VO4 |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:6.10 Å Release Date: 2016-09-21 Classification: LYASE |
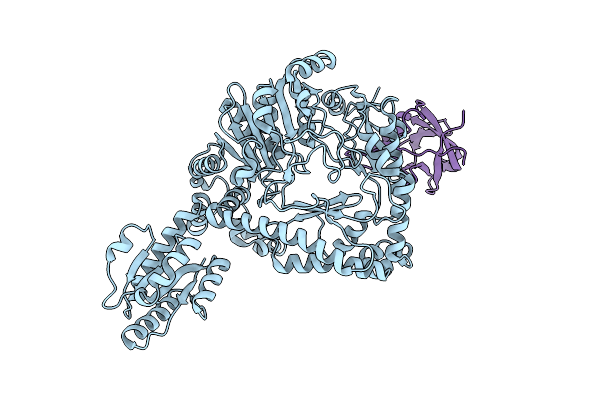 |
Revisited Cryo-Em Structure Of Inducible Lysine Decarboxylase Complexed With Lara Domain Of Rava Atpase
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:6.20 Å Release Date: 2016-09-21 Classification: HYDROLASE/ISOMERASE |
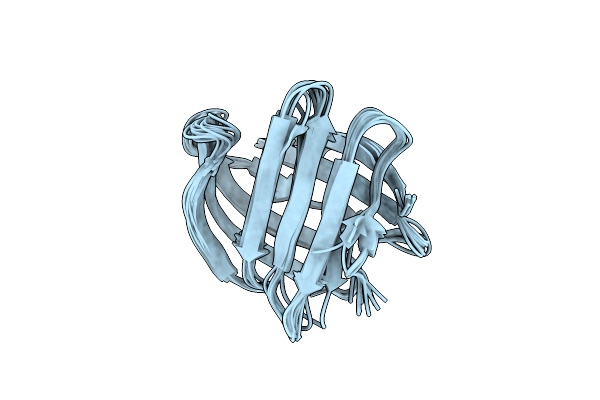 |
The Nmr Structure Of A Stable And Compact All-Beta-Sheet Variant Of Intestinal Fatty Acid-Binding Protein
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 2004-06-08 Classification: LIPID BINDING PROTEIN |
 |
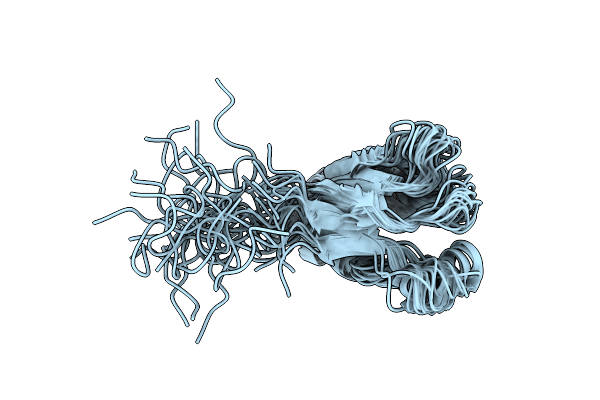
The Three-Dimensional Structure Of A Helix-Less Variant Of Intestinal Fatty Acid Binding Protein, Nmr, 20 Structures
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 1998-05-27 Classification: FATTY ACID-BINDING |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1997-09-26 Classification: NUCLEAR PROTEIN |

