Search Count: 855
 |
Organism: Escherichia coli o139:h28 (strain e24377a / etec), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: 4NE |
 |
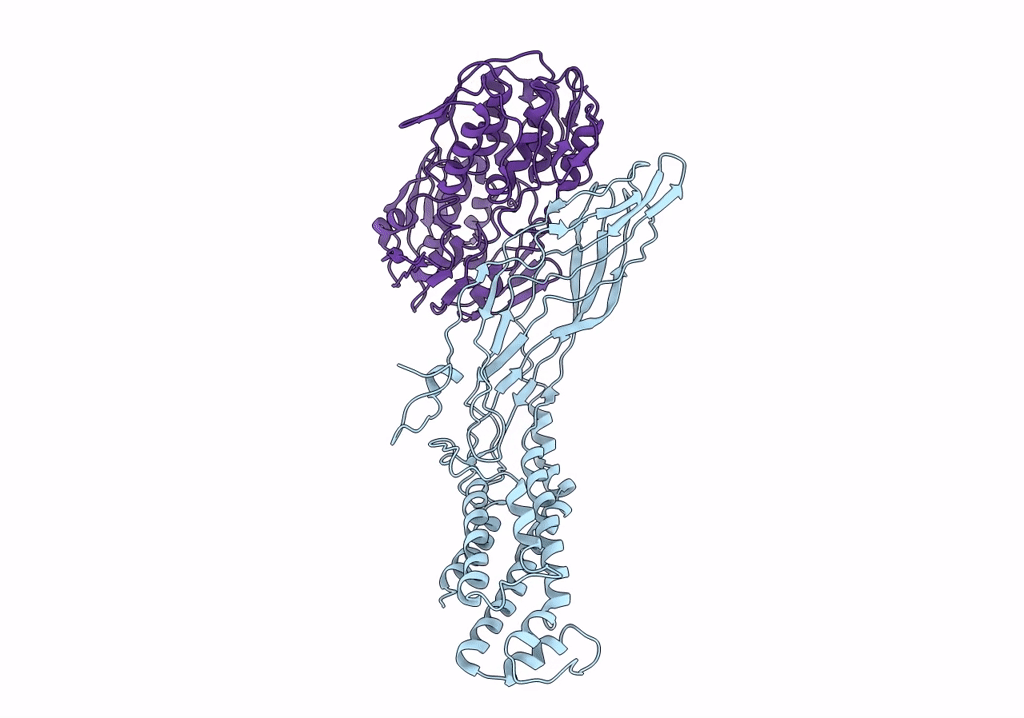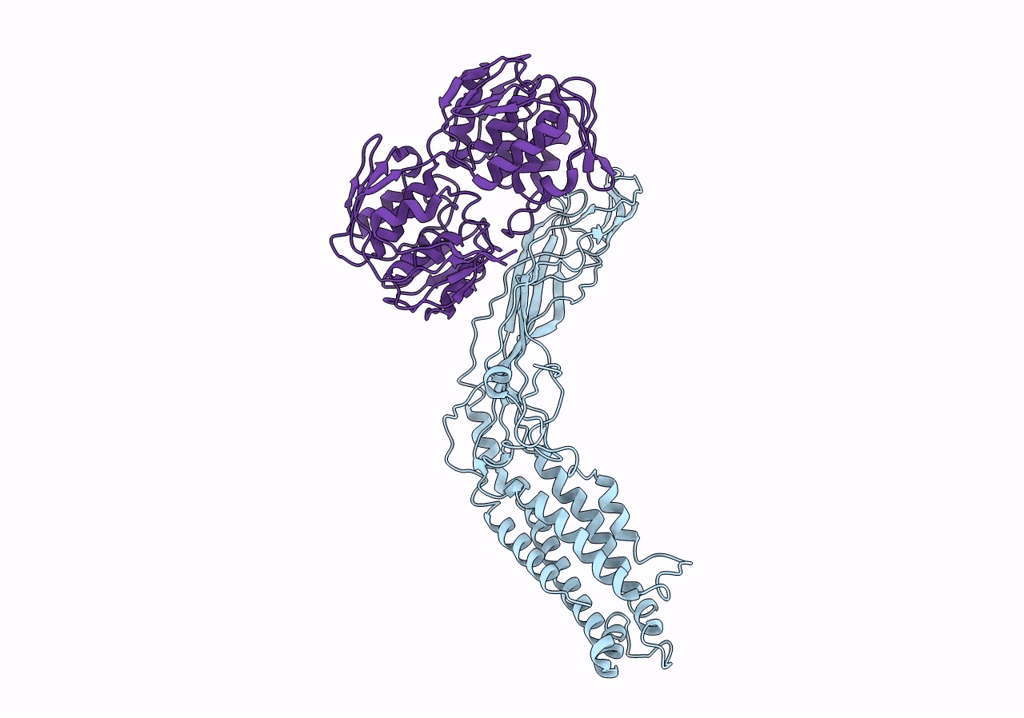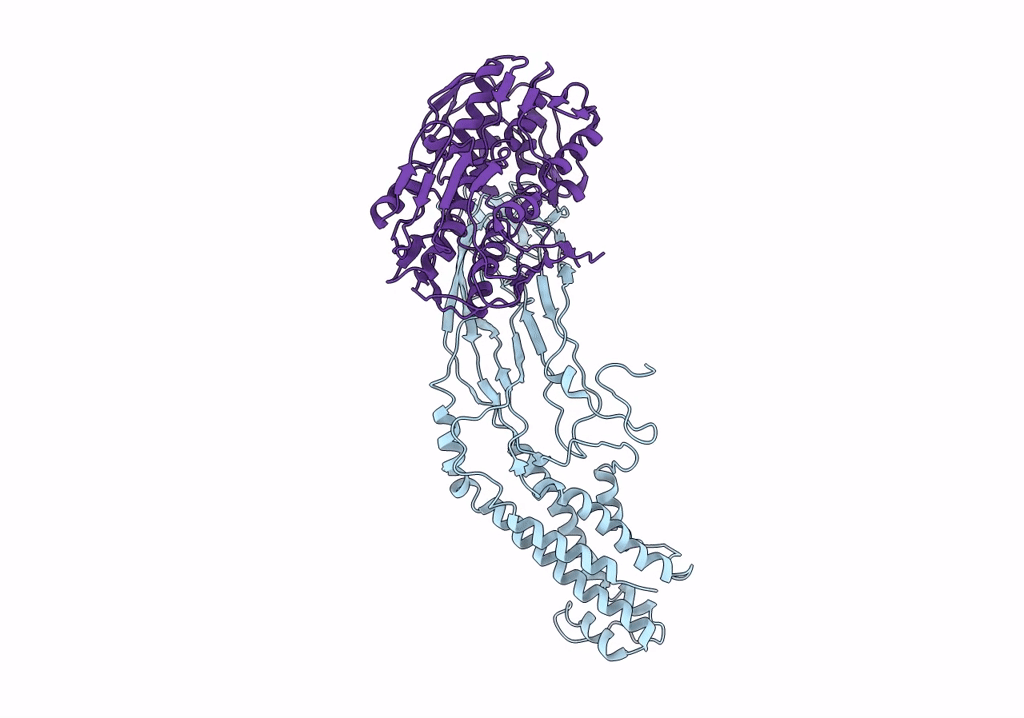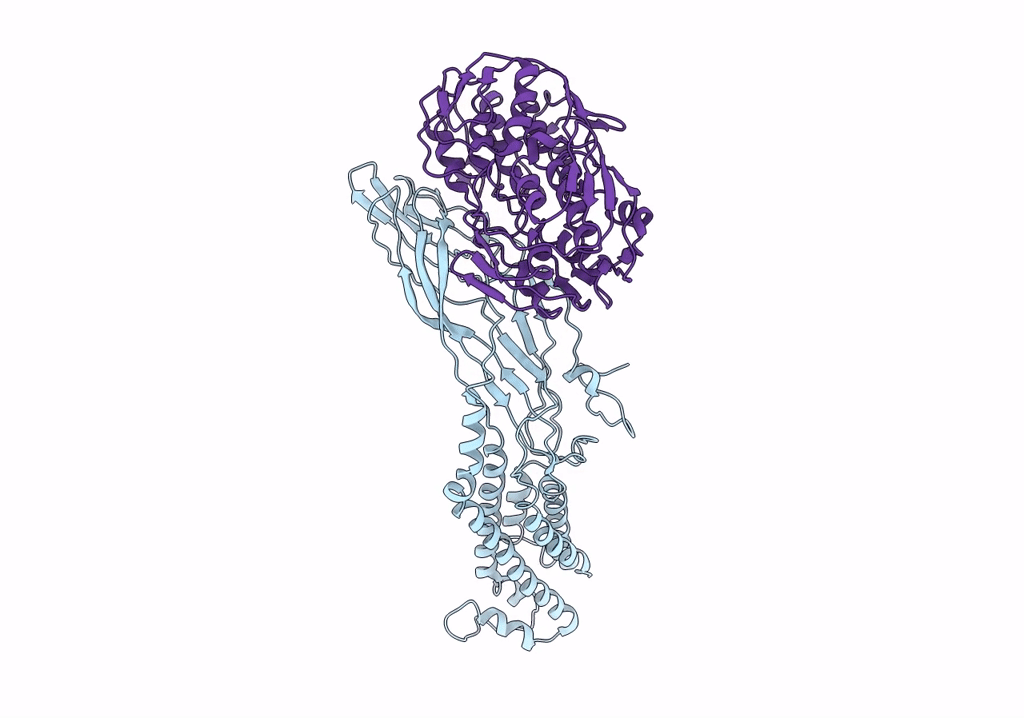
Crystal Structure Of A Peptidergic Gpcr In Complex With A Small Synthetic G Protein-Biased Agonist
Organism: Homo sapiens, Escherichia coli o139:h28 str. e24377a
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-12-04 Classification: MEMBRANE PROTEIN Ligands: A1D5N, OLA, 1PE |
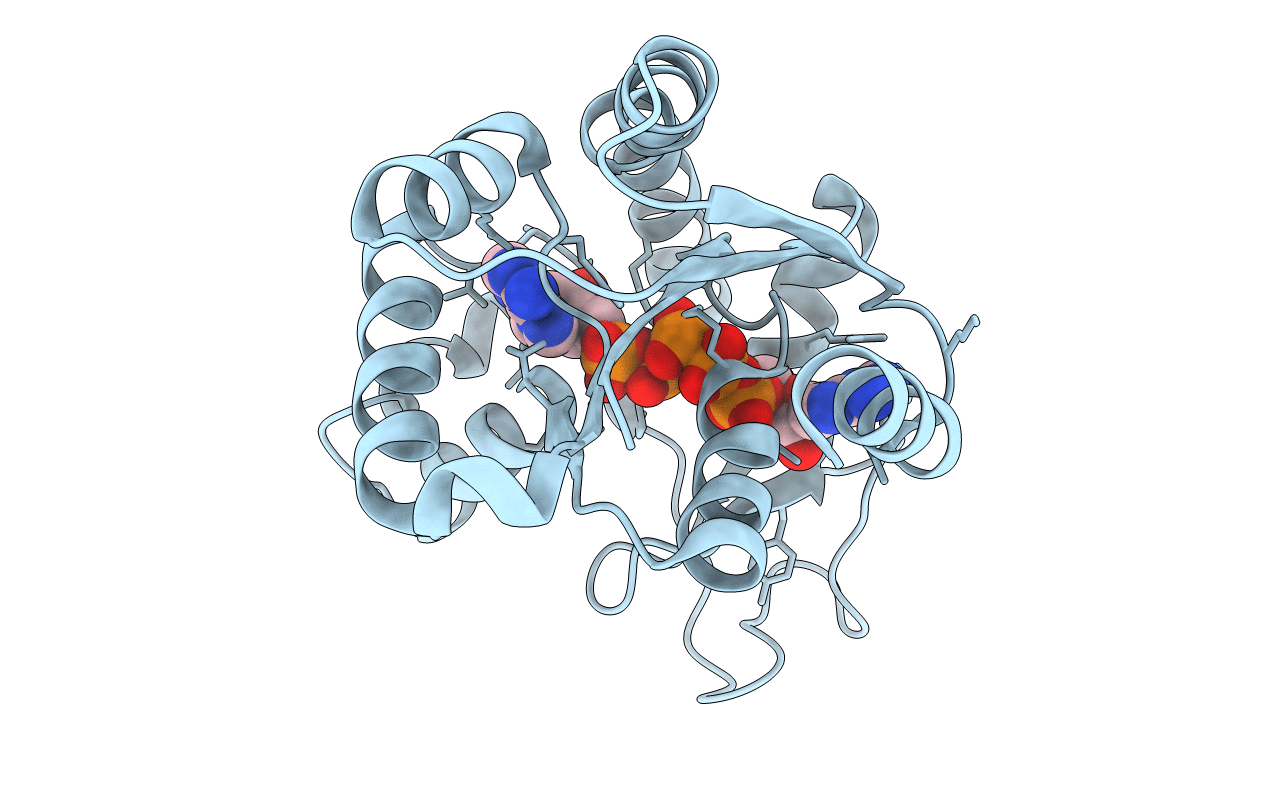 |
Organism: Escherichia coli o139:h28 (strain e24377a / etec)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-08-28 Classification: TRANSFERASE Ligands: AP5 |
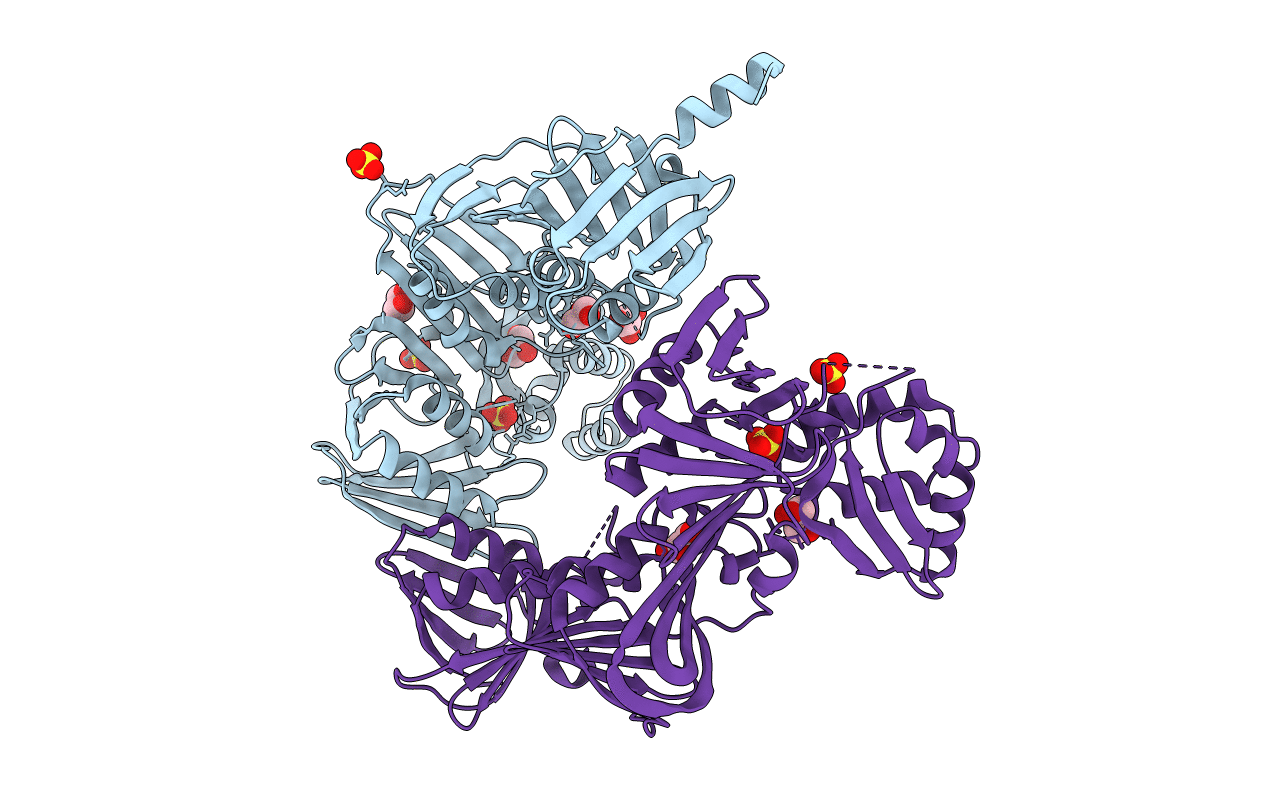 |
Organism: Escherichia coli (strain k12), Escherichia coli o139:h28 (strain e24377a / etec)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-05-01 Classification: DNA BINDING PROTEIN Ligands: GOL, SO4 |
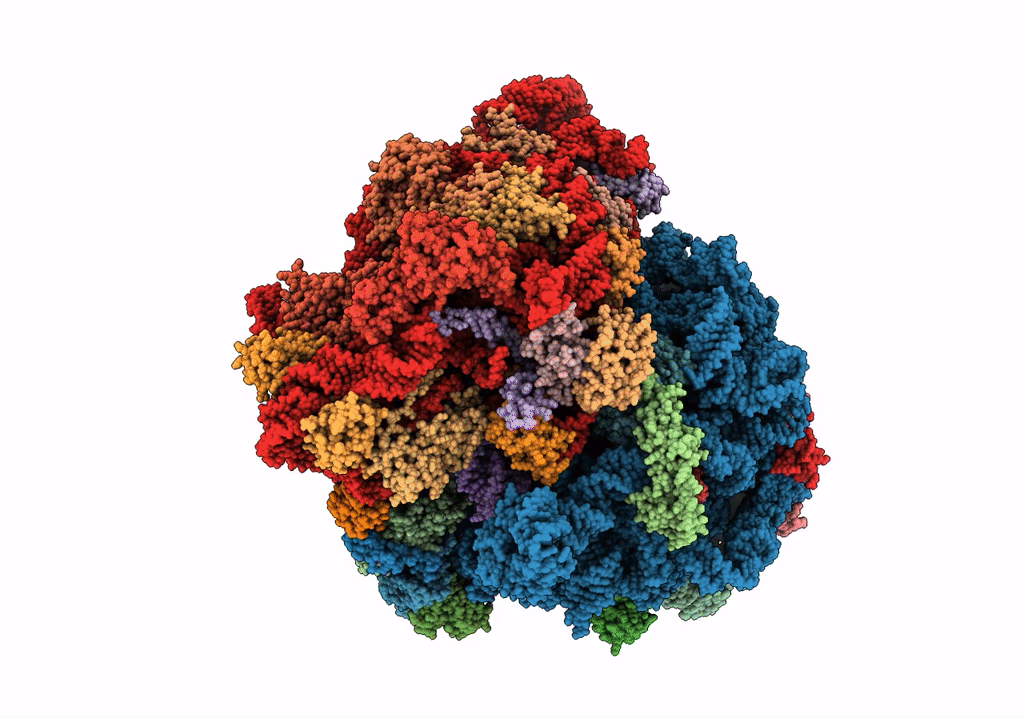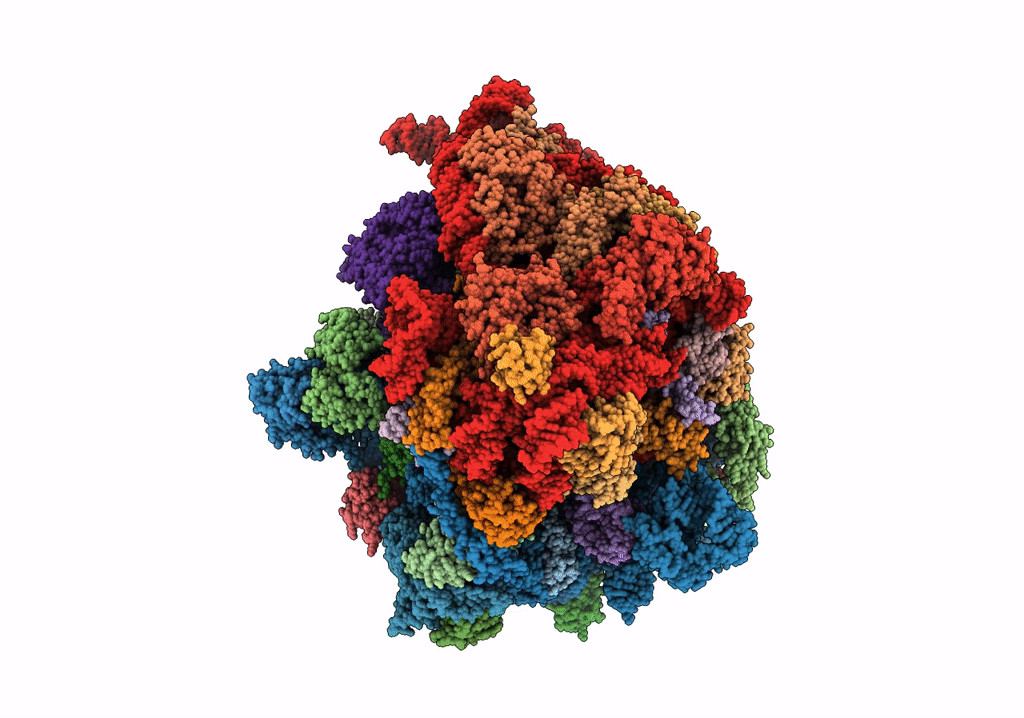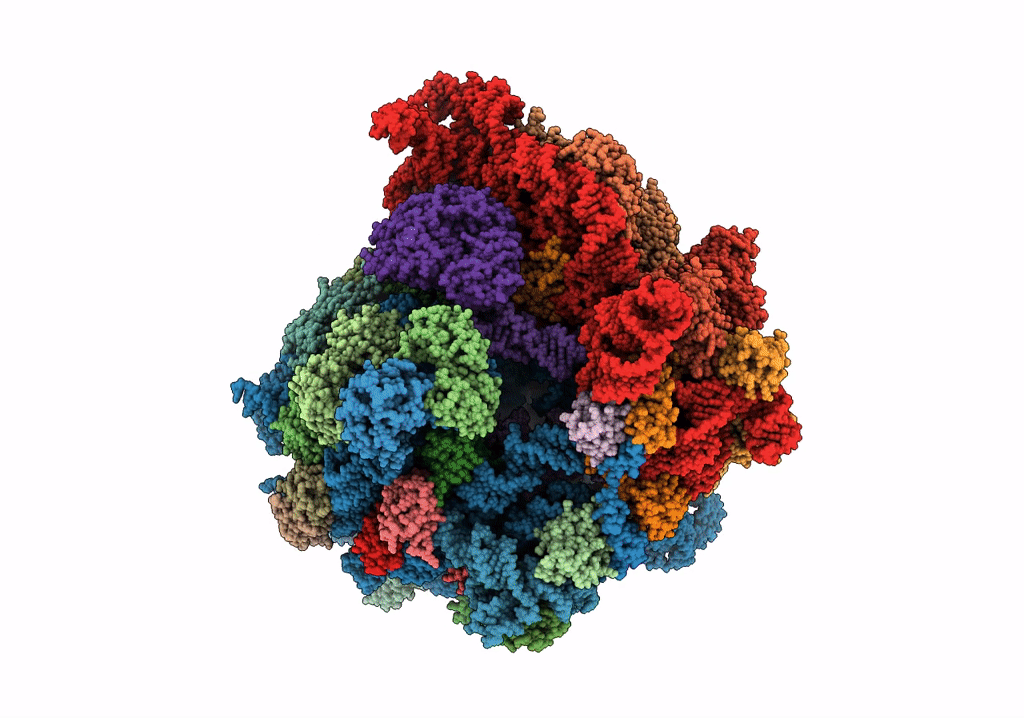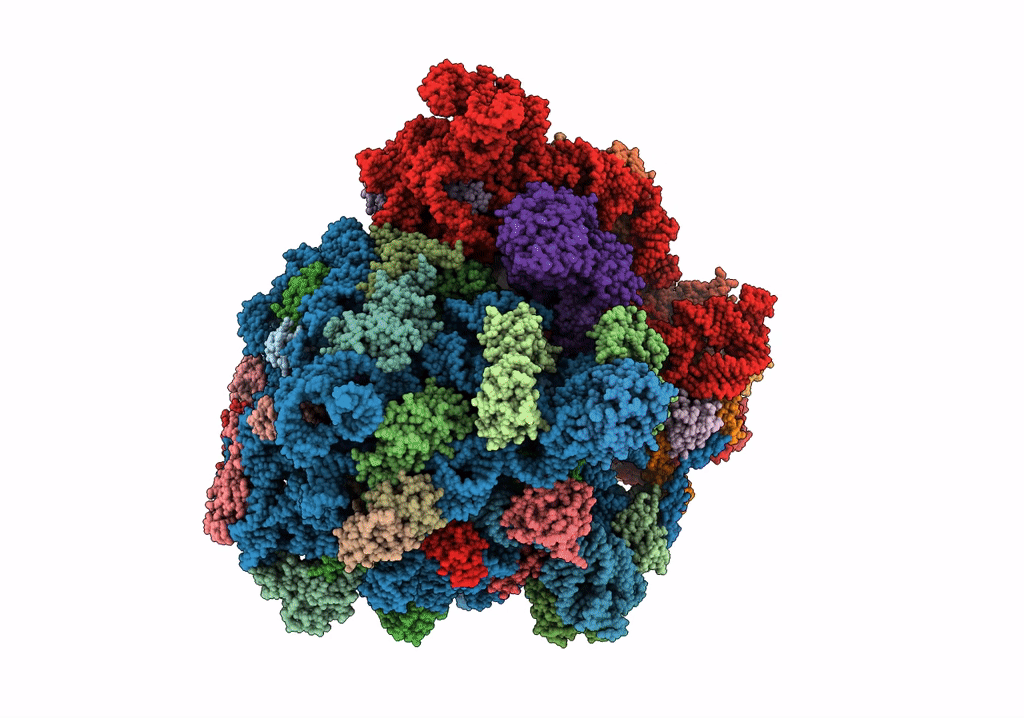 |
Organism: Escherichia coli, Escherichia coli o139:h28 (strain e24377a / etec)
Method: ELECTRON MICROSCOPY Release Date: 2018-05-02 Classification: RIBOSOME Ligands: FME, MG, K, PHE, GTP |
 |
Organism: Escherichia phage qbeta, Escherichia coli o139:h28 (strain e24377a / etec)
Method: ELECTRON MICROSCOPY Release Date: 2017-10-18 Classification: VIRUS/TRANSFERASE |
 |
Crystal Structure Of E.Coli Purine Nucleoside Phosphorylase With Acycloguanosine
Organism: Escherichia coli o139:h28 (strain e24377a / etec)
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2017-02-22 Classification: TRANSFERASE Ligands: AC2, SO4 |
 |
Organism: Escherichia coli o139:h28, Synthetic construct, Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Escherichia coli jj1887
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2016-10-12 Classification: RIBOSOME Ligands: MG, ZN, SF4 |
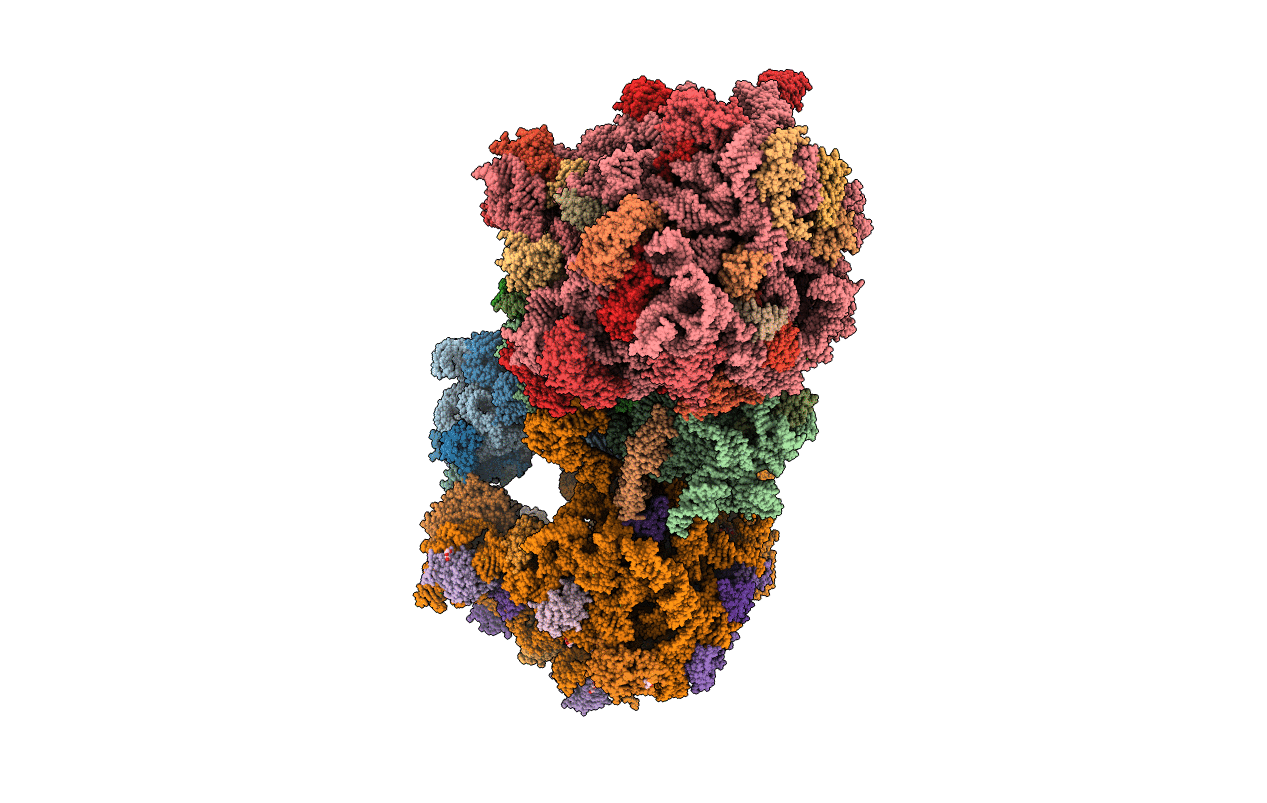 |
Structure Of The E Coli 70S Ribosome With The U1052G Mutation In 16S Rrna Bound To Tigecycline
Organism: Escherichia coli, Escherichia coli (strain k12), Escherichia coli o139:h28 (strain e24377a / etec)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-07-06 Classification: ribosome/antibiotic Ligands: MG, PG4, MPD, PUT, T1C, ZN, PEG, EDO, PGE, SPD, 1PE, ACY, GUN, TRS |
 |
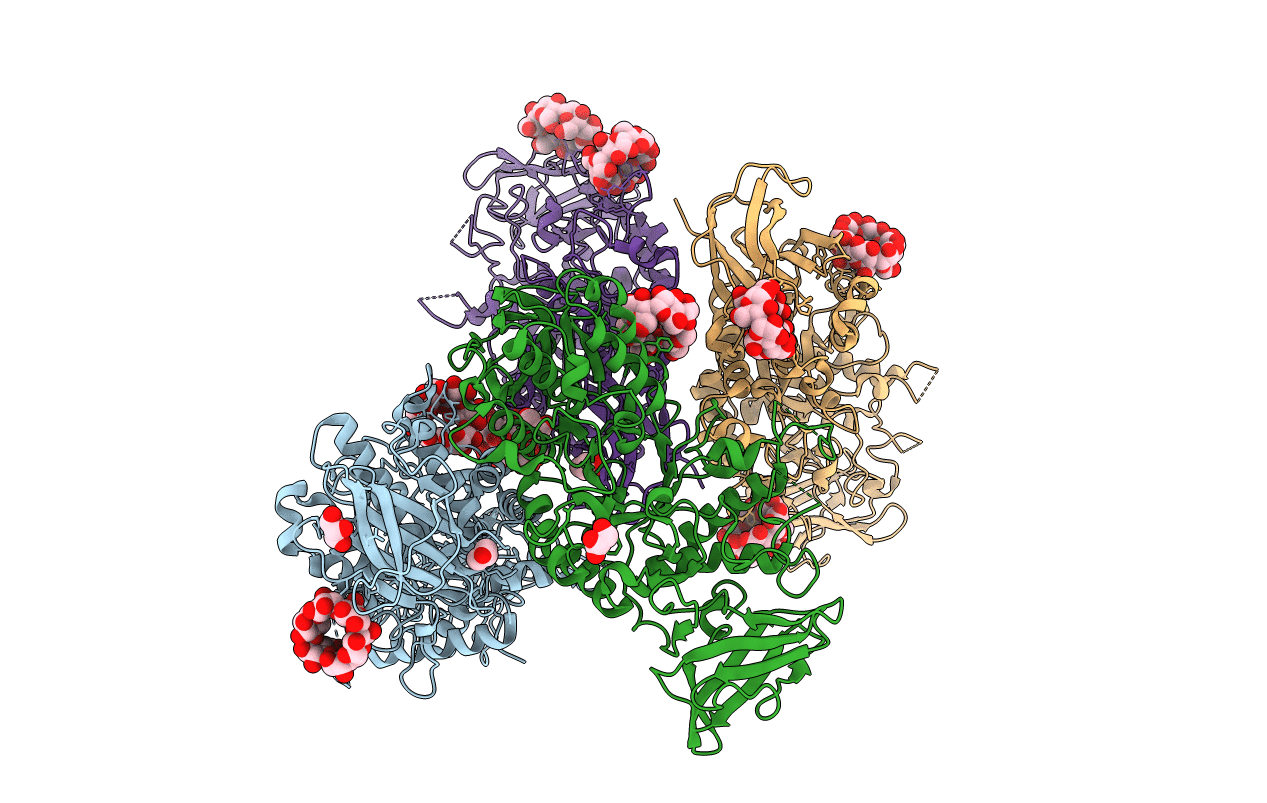
Structure Of The K. Pneumonia Slma-Dna Complex Bound To The C-Terminal Of The Cell Division Protein Ftsz
Organism: Escherichia coli o139:h28 (strain e24377a / etec), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2016-06-22 Classification: DNA BINDING PROTEIN/DNA |
 |
Crystal Structure Of E.Coli Branching Enzyme In Complex With Alpha Cyclodextrin
Organism: Escherichia coli o139:h28 (strain e24377a / etec)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-12-16 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Escherichia coli o139:h28 (strain e24377a / etec)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2015-12-16 Classification: TRANSFERASE Ligands: GOL |
 |
Organism: Escherichia coli o139:h28 (strain e24377a / etec)
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2015-12-16 Classification: TRANSFERASE Ligands: GOL |
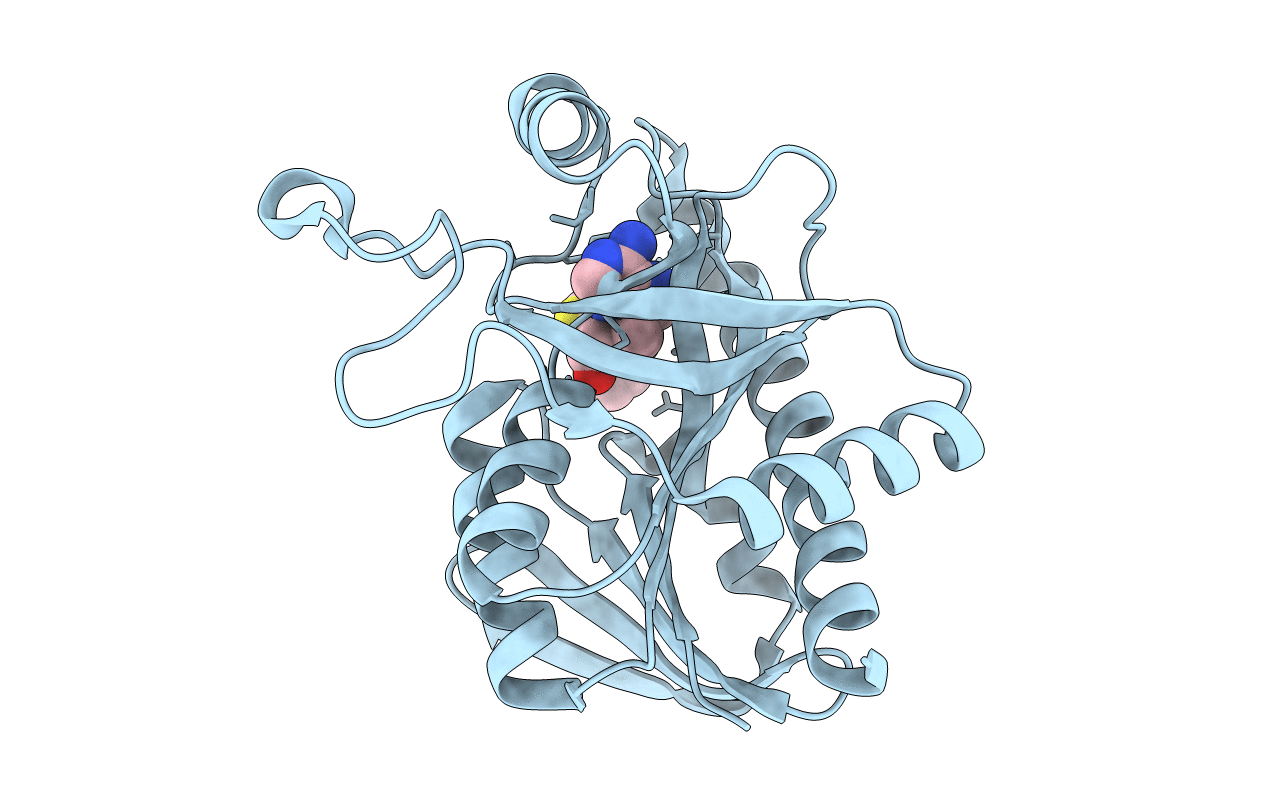 |
Crystal Structure Of Escherichia Coli 5'-Methylthioadenosine/S-Adenosyl Homocysteine Nucleosidase (Mtan) Complexed With Butylthio-Dadme-Immucillin-A
Organism: Escherichia coli o139:h28
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2015-08-19 Classification: hydrolase/hydrolase inhibitor Ligands: BIG, PG4 |
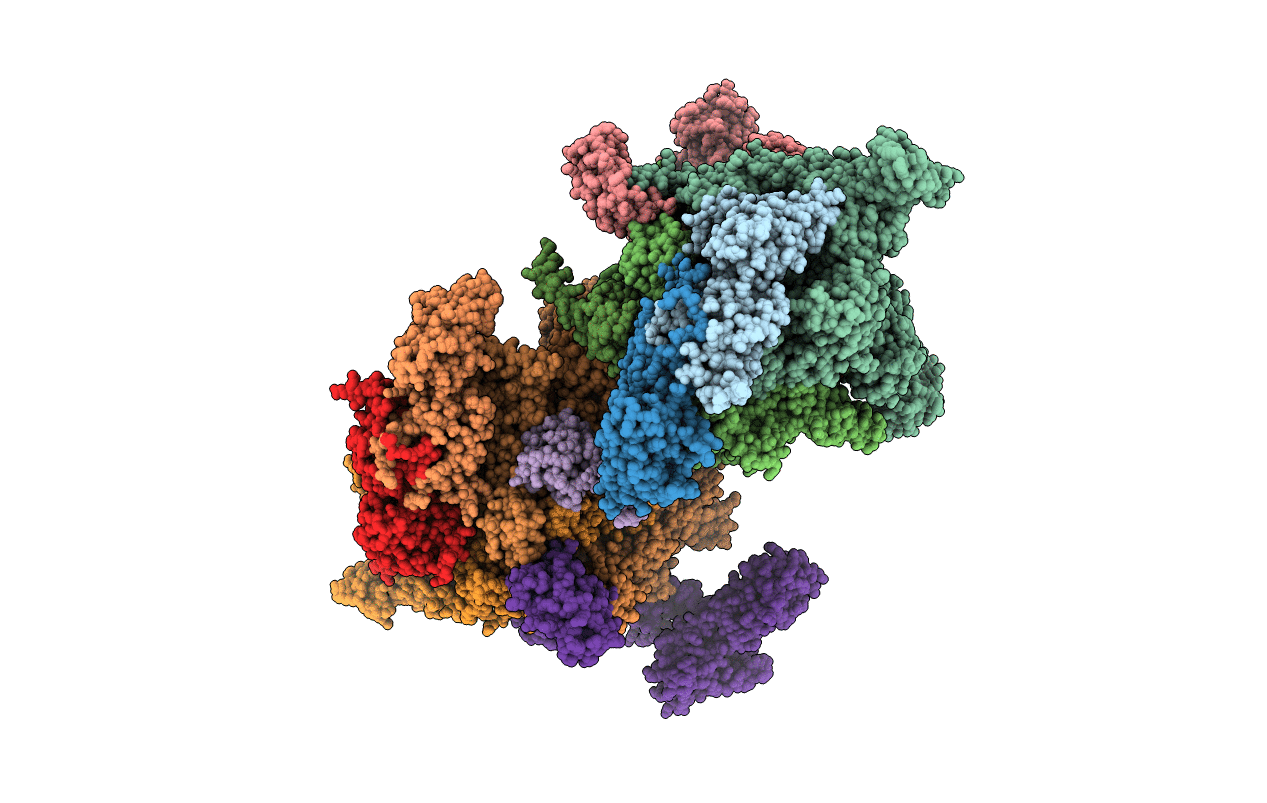 |
Crystal Structure Of Cbr 703 Bound To Escherichia Coli Rna Polymerase Holoenzyme
Organism: Escherichia coli (strain k12), Escherichia coli o139:h28 (strain e24377a / etec), Citrobacter koseri (strain atcc baa-895 / cdc 4225-83 / sgsc4696)
Method: X-RAY DIFFRACTION Resolution:3.71 Å Release Date: 2015-07-22 Classification: transcription/antibiotic Ligands: 42S, MG, ZN |
 |
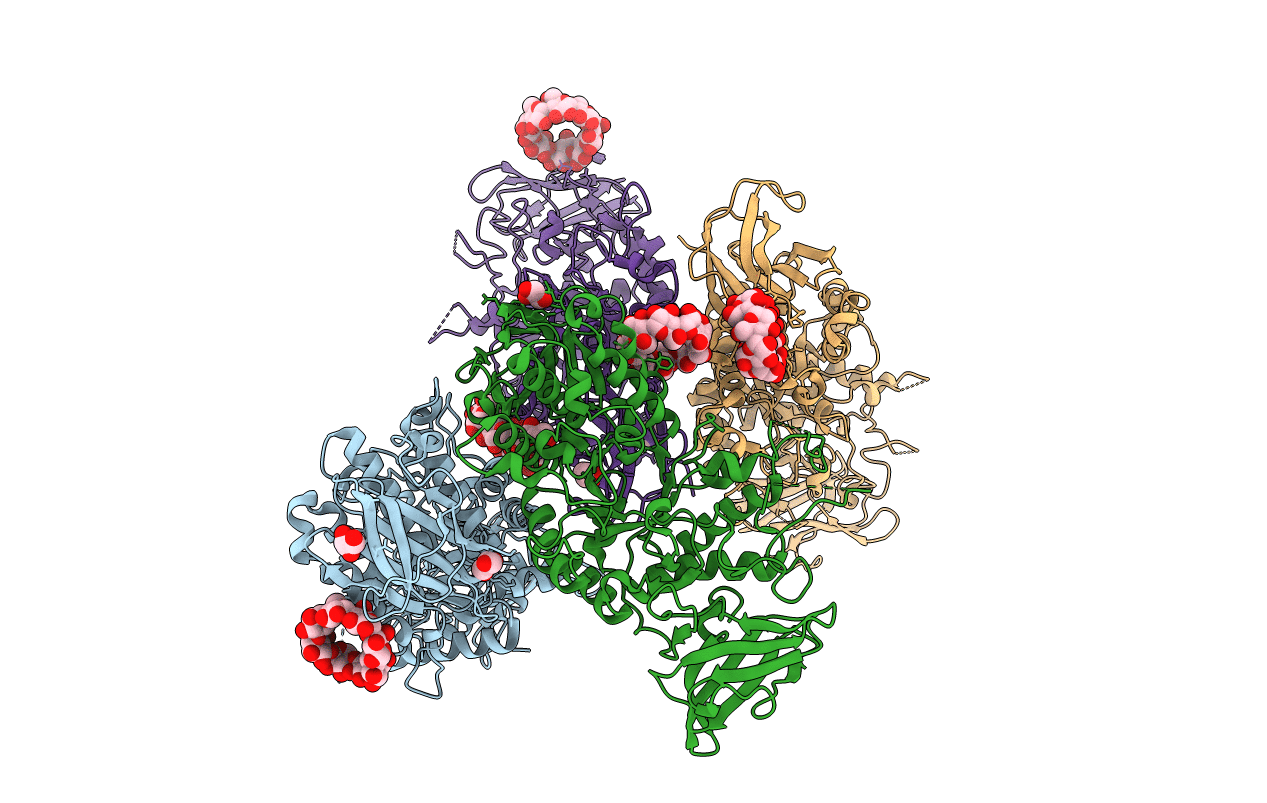
Crystal Structure Of Cbr 9379 Bound To Escherichia Coli Rna Polymerase Holoenzyme
Organism: Escherichia coli (strain k12), Escherichia coli o139:h28 (strain e24377a / etec), Citrobacter koseri (strain atcc baa-895 / cdc 4225-83 / sgsc4696)
Method: X-RAY DIFFRACTION Resolution:4.01 Å Release Date: 2015-07-22 Classification: transcription/antibiotic Ligands: 42T, MG, ZN |
 |
Crystal Structure Of Cbr 9393 Bound To Escherichia Coli Rna Polymerase Holoenzyme
Organism: Escherichia coli (strain k12), Escherichia coli o139:h28 (strain e24377a / etec), Escherichia coli (strain atcc 8739 / dsm 1576 / crooks)
Method: X-RAY DIFFRACTION Resolution:3.68 Å Release Date: 2015-07-22 Classification: transcription/antibiotic Ligands: 42U, MG, ZN |
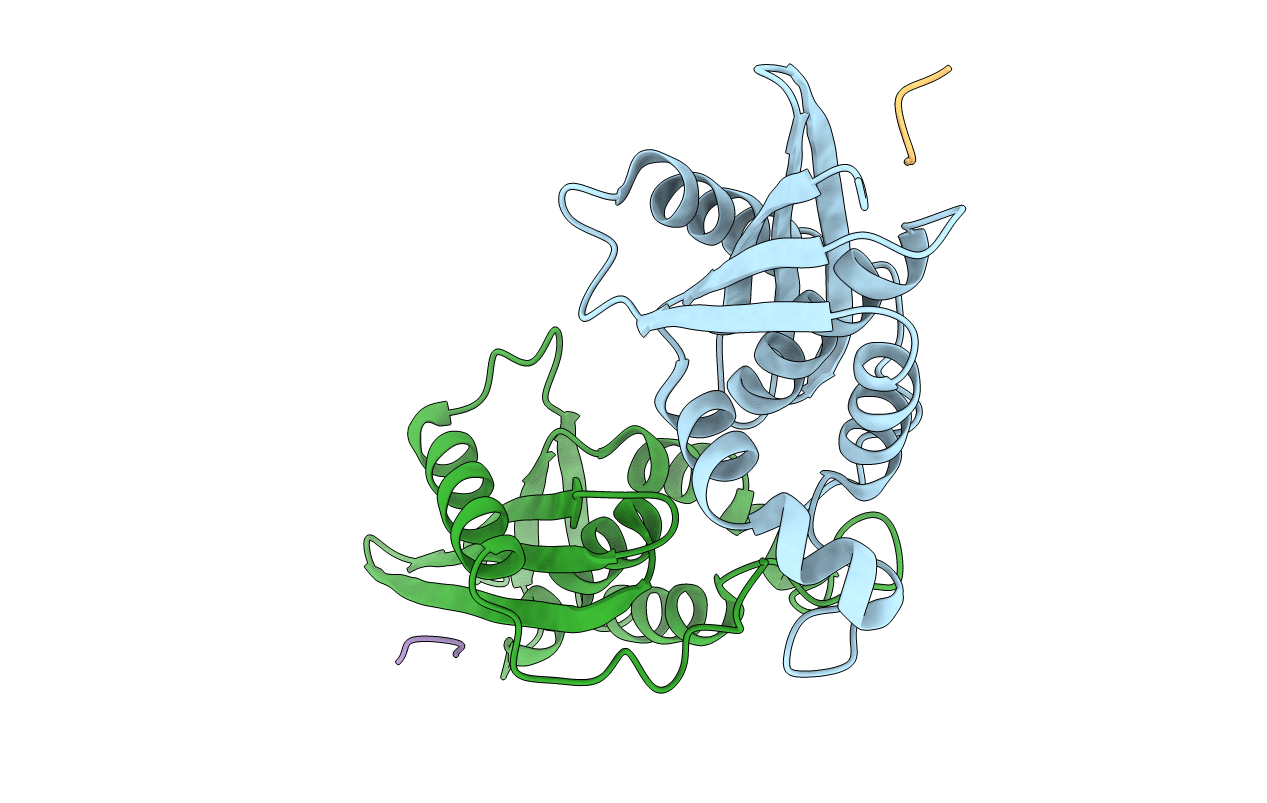 |
Organism: Escherichia coli o139:h28, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-04-29 Classification: Hydrolase/Peptide |
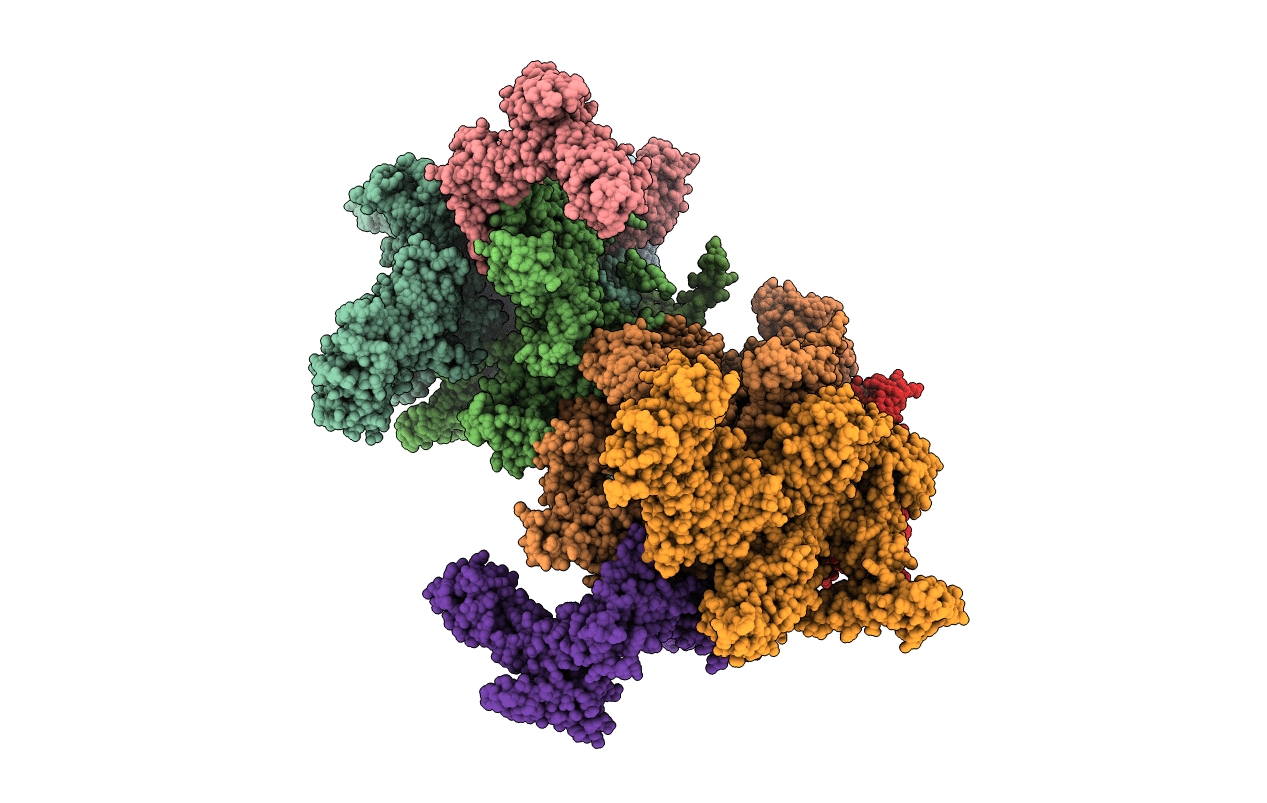 |
Organism: Escherichia coli o139:h28 (strain e24377a / etec), Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.57 Å Release Date: 2015-03-11 Classification: transcription/transcription inhibitor Ligands: MG, ZN, 4C6 |
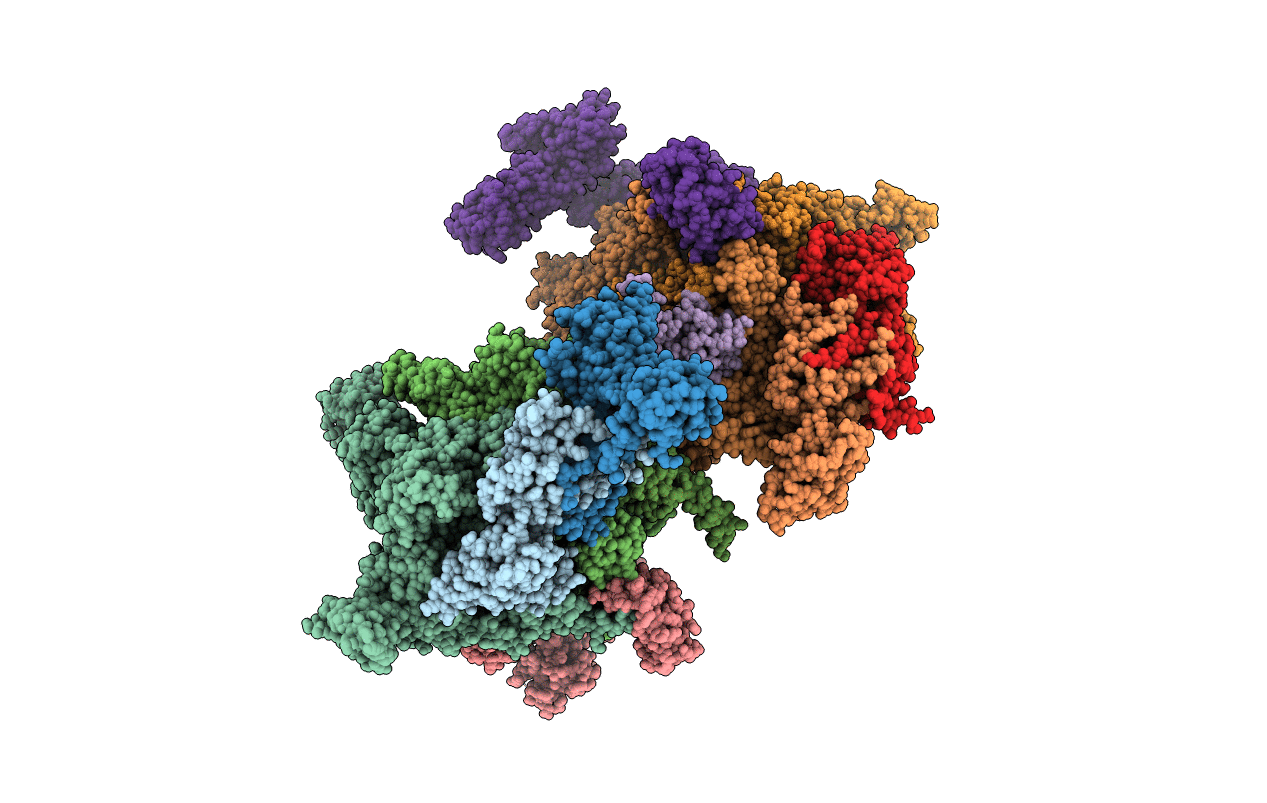 |
Escherichia Coli Rna Polymerase In Complex With Squaramide Compound 14 (N-[3,4-Dioxo-2-(4-{[4-(Trifluoromethyl)Benzyl]Amino}Piperidin-1-Yl)Cyclobut-1-En-1-Yl]-3,5-Dimethyl-1,2-Oxazole-4-Sulfonamide)
Organism: Escherichia coli o139:h28 (strain e24377a / etec), Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.82 Å Release Date: 2015-03-11 Classification: transcription/transcription inhibitor Ligands: MG, ZN, 4C2 |

