Search Count: 298
 |
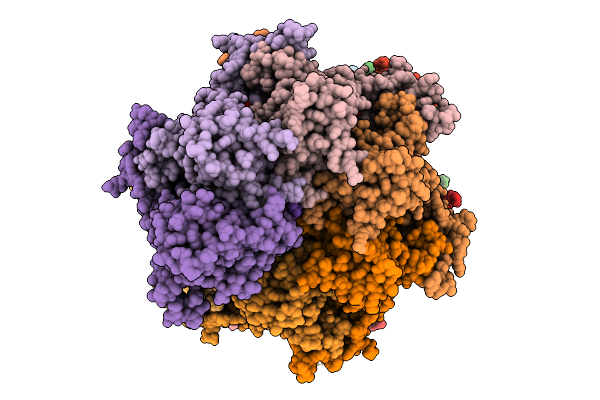
Sixteen Polymer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Twenty-Two Polymer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: MG, ATP |
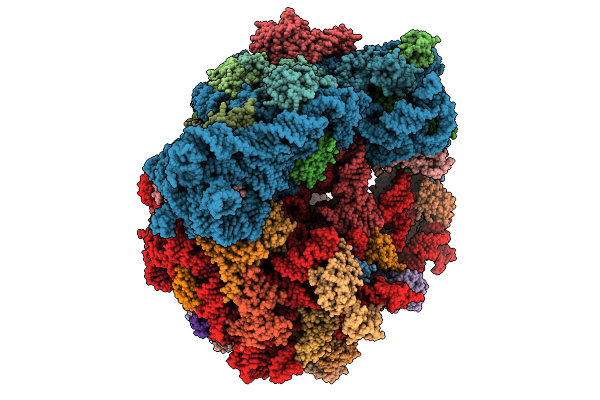 |
Organism: Escherichia coli, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: RIBOSOME Ligands: ZN, K, MG, PRO |
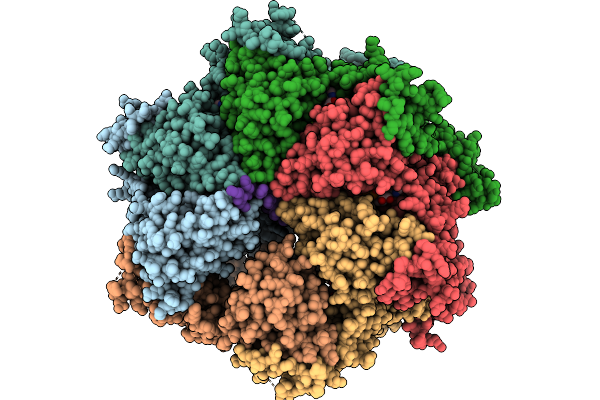 |
Organism: Escherichia coli bl21(de3), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: TRANSLOCASE Ligands: ATP, MG |
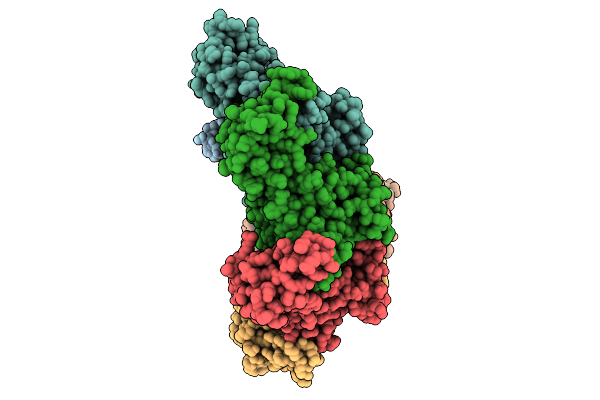 |
Hexamer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: MG, ATP |
 |
Hexamer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Resolution:2.87 Å Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Hexamer Msp1 From S.Cerevisiae(With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-12-31 Classification: HYDROLASE Ligands: ZN, A1EJI, GOL |
 |
Hexamer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Escherichia coli bl21(de3), Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: SFG, ZN, SO4 |
 |
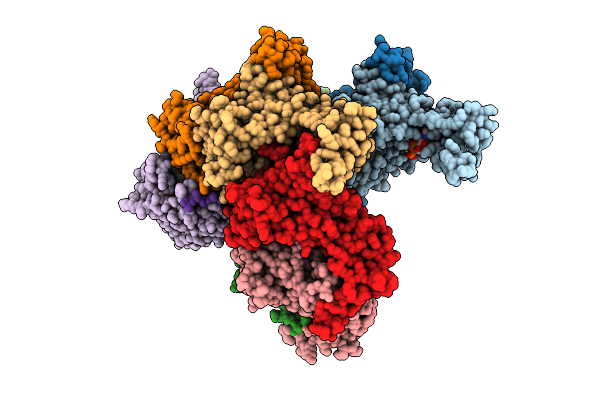
Cryo-Em Structure Of Ctpa S300A/K325A/Q329A Mutant From Helicobacter Pylori
Organism: Helicobacter pylori g27, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Resolution:3.14 Å Release Date: 2025-12-03 Classification: HYDROLASE |
 |
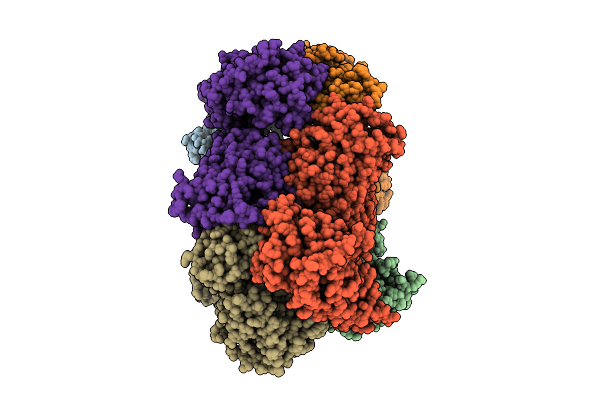
Nanomer Msp1 From S.Cerevisiae (With A Catalytic Dead Mutation) In Complex With An Unknown Peptide Substrate
Organism: Saccharomyces cerevisiae s288c, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
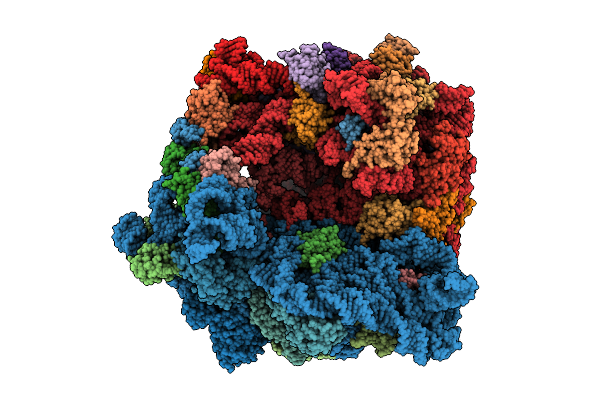
Organism: Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Resolution:2.89 Å Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
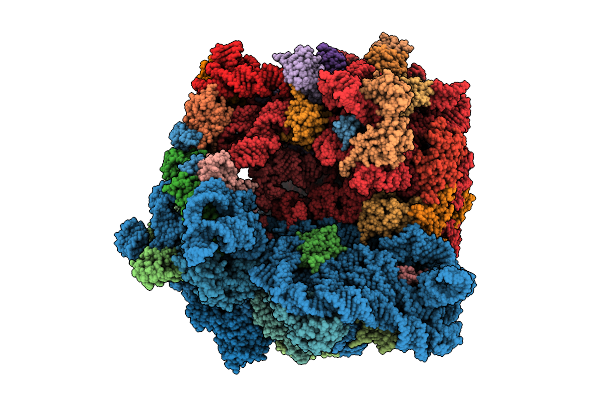
Structure Of The Bacterial Ribosome Without Hypoxia-Induced Rrna Modifications
Organism: Escherichia phage t4, Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG |
 |
Structure Of The Bacterial Ribosome With Hypoxia-Induced Rrna Modifications
Organism: Escherichia phage t4, Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RIBOSOME Ligands: MG |
 |
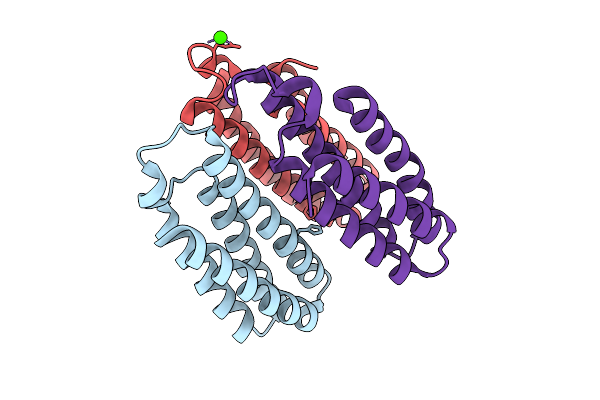
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: CO, CA, CL, NA |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: NI, CL, CA |
 |
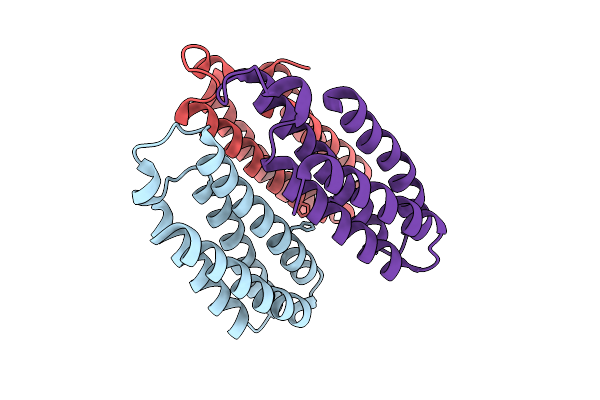
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: ZN, CA, NA |

