Search Count: 15
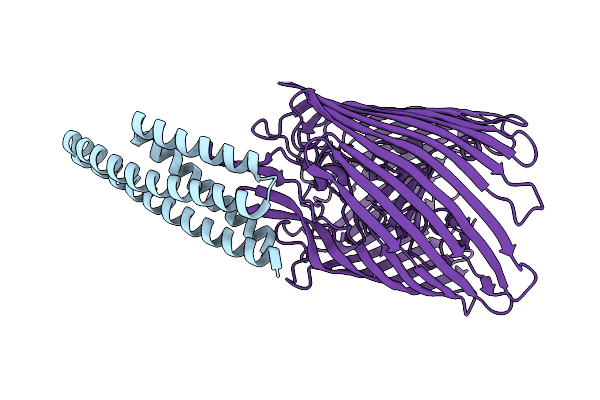 |
Cryo-Em Structure Of The Heme/Hemoglobin Transporter Chua, In Complex With De Novo Designed Binder G7
Organism: Synthetic construct, Escherichia coli cft073
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSPORT PROTEIN |
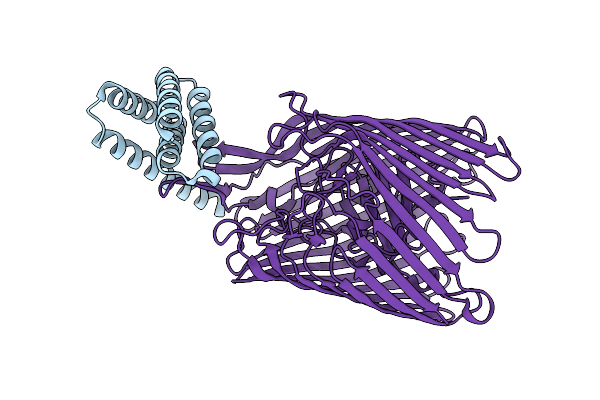 |
Cryo-Em Structure Of The Heme/Hemoglobin Transporter Chua, In Complex With De Novo Designed Binder H3
Organism: Synthetic construct, Escherichia coli cft073
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSPORT PROTEIN |
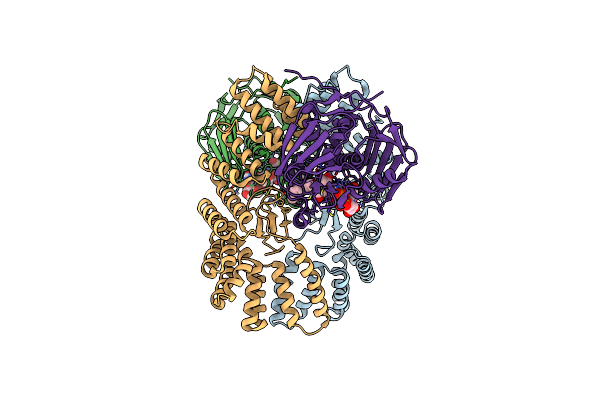 |
Organism: Escherichia coli cft073
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: PROTEIN BINDING Ligands: ZN, 24G, ACT |
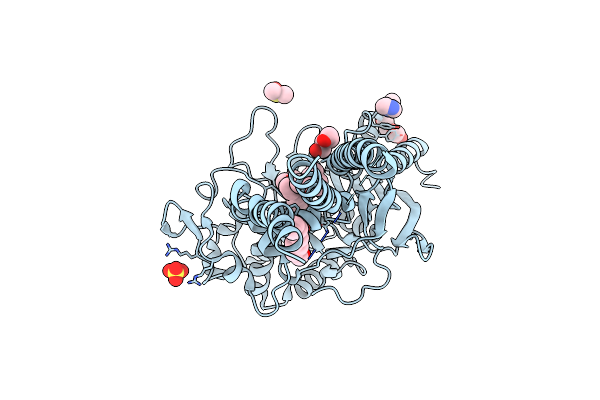 |
Organism: Escherichia coli cft073
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-09-28 Classification: HYDROLASE Ligands: OLC, 2PE, IMD, DMS, SO4, CL |
 |
Organism: Escherichia coli cft073
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-09-28 Classification: HYDROLASE/INHIBITOR Ligands: DMS, Z9A, Z9G, 2PE, AV0, 97N, SO4, CL |
 |
Organism: Escherichia coli cft073
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: HYDROLASE |
 |
Organism: Escherichia coli cft073
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: MEMBRANE PROTEIN Ligands: 6OU |
 |
Organism: Escherichia coli cft073
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
Organism: Escherichia coli cft073, Escherichia coli k12
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2019-02-27 Classification: OXIDOREDUCTASE Ligands: SF4, F3S, SF3, LMT, CL, FCO, NI, MG, LI, SO4 |
 |
Organism: Escherichia coli cft073
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2016-03-09 Classification: UNKNOWN FUNCTION Ligands: CL, FMT, NA, EDO |
 |
Bacterial Arylsulfate Sulfotransferase (Asst) H436N Mutant With 4-Nitrophenyl Sulfate (Pns) In The Active Site
Organism: Escherichia coli cft073
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-03-26 Classification: TRANSFERASE Ligands: 4NS, SO4 |
 |
Bacterial Arylsulfate Sulfotransferase (Asst) H436N Mutant With 4-Methylumbelliferyl Sulfate (Mus) In The Active Site
Organism: Escherichia coli cft073
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-03-26 Classification: TRANSFERASE Ligands: SO4, MUX |
 |
Organism: Escherichia coli cft073
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-03-26 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Escherichia coli cft073
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2010-08-18 Classification: HYDROLASE INHIBITOR Ligands: BME, MPD, SO4, CL |
 |
Crystal Structure Of The Putative Capsid Protein Of Prophage (E.Coli Cft073)
Organism: Escherichia coli cft073
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-01-08 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |

