Search Count: 41,455
 |
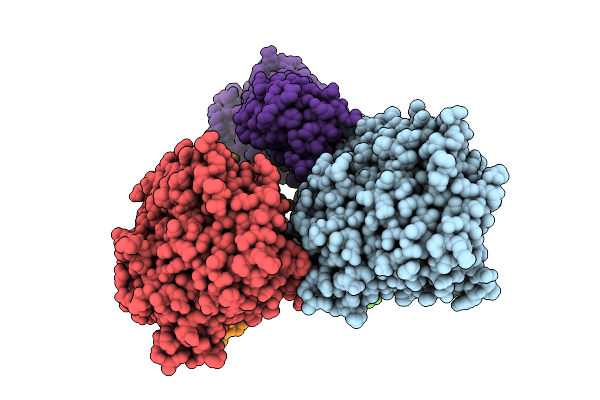
The Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Compound Zm_097
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: A1D76, DMS |
 |
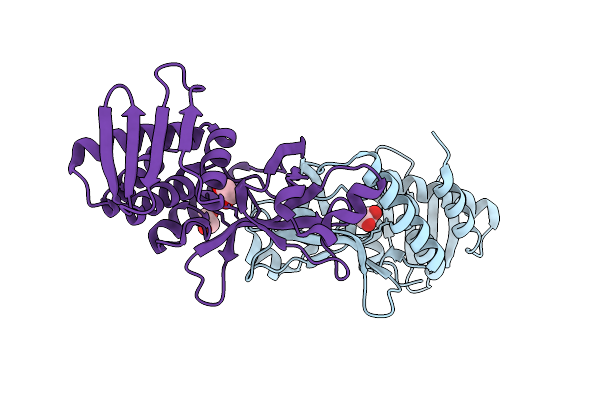
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CELL CYCLE Ligands: G2P, MG, GTP |
 |
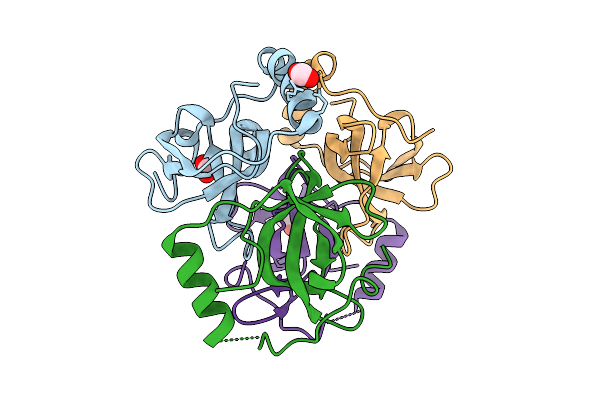
Structure Of The Tela-Associated Type Vii Secretion System Chaperone Sir_0168
Organism: Streptococcus intermedius b196
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: EDO, CL |
 |
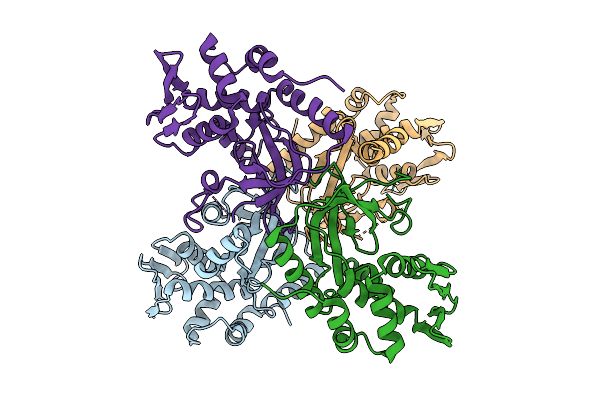
Organism: Streptococcus intermedius
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: FMT, ACY |
 |
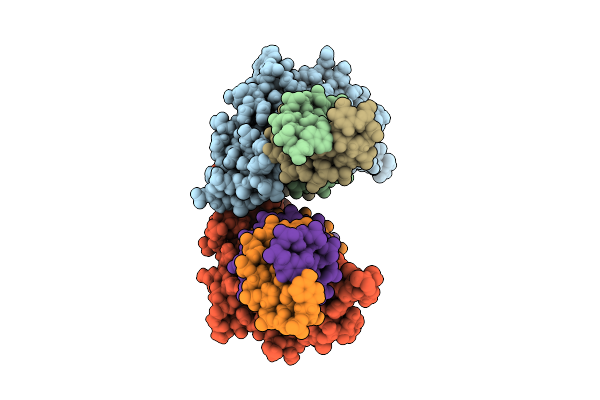
Structure Of The Telb-Associated Type Vii Secretion System Chaperone Sir_0178
Organism: Streptococcus intermedius b196
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: CHAPERONE Ligands: CL |
 |
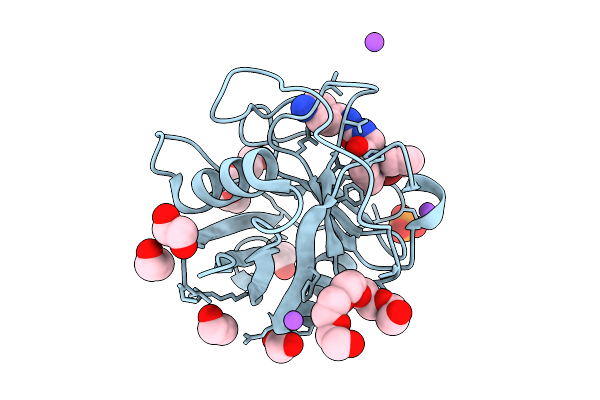
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
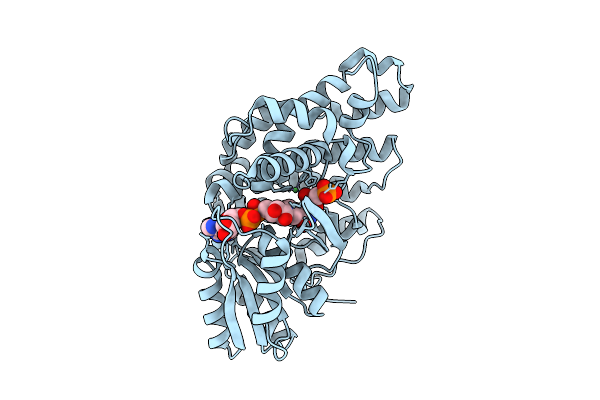
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: 9CK, EDO, 1PE, PGE, PO4, NA |
 |
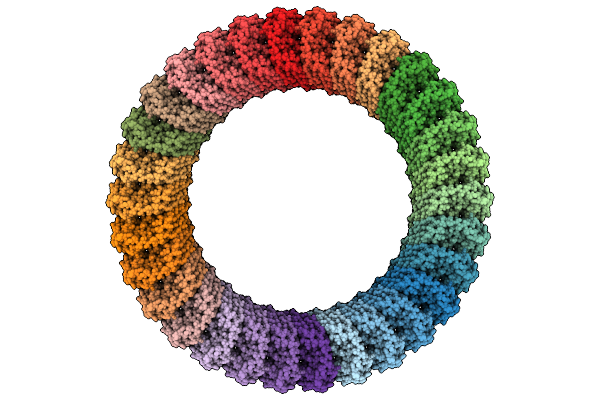
Crystal Structure Of Ni2+ Dependent Glycerol-1-Phosphate Dehydrogenase Aram From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: 13P, NAI, NI |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: APOPTOSIS |
 |
X-Ray Structure Of Trichomonas Vaginalis Inactive Mutant Hydrogenosomal Processing Peptidase Heterodimer (Hppin)
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: SO4, ZN |
 |
X-Ray Structure Of Hydrogenosomal Processing Peptidase (Hpp), E56Q Inactive Mutant, From Trichomonas Vaginalis Co-Crystallized With Presequence Peptide From Adenylate Kinase (Ak) - Not Visible In The Structure Model
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: SO4, GOL, ZN |
 |
Bacteroides Ovatus Gh98 Endoxylanase In Complex With Arabino-Xylooligosaccharide
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: AHR, CA, XYP |
 |
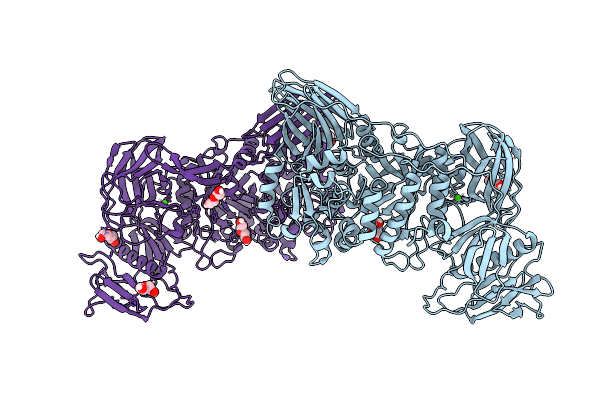
Inactive Bacteroides Ovatus Gh98 Endoxylanase E361A In Complex With Arabino-Xylooligosaccharide
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: CA |
 |
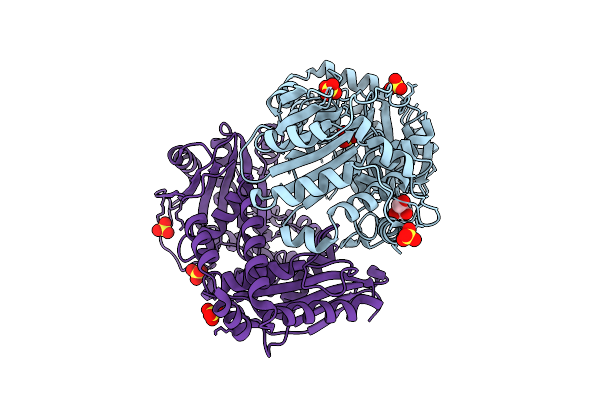
Organism: Bacteroides ovatus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: PEG, CA |
 |
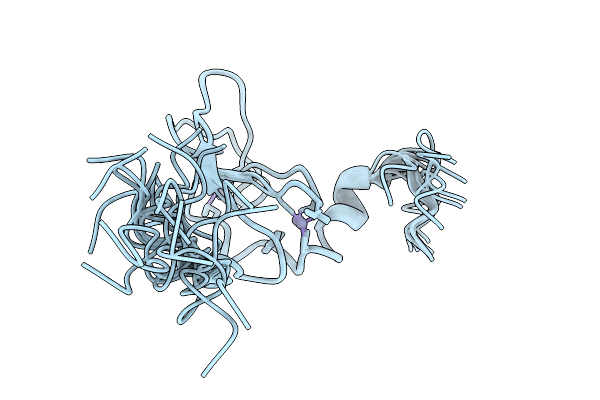
X-Ray Structure Of Hydrogenosomal Processing Peptidase (Hpp), E56Q Inactive Mutant, From Trichomonas Vaginalis Co-Crystallized With Presequence Peptide From Ferredoxin Oxidoreductase (Pfo) - Not Visible In The Structure Model
Organism: Trichomonas vaginalis
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: SO4, GOL, ZN |
 |
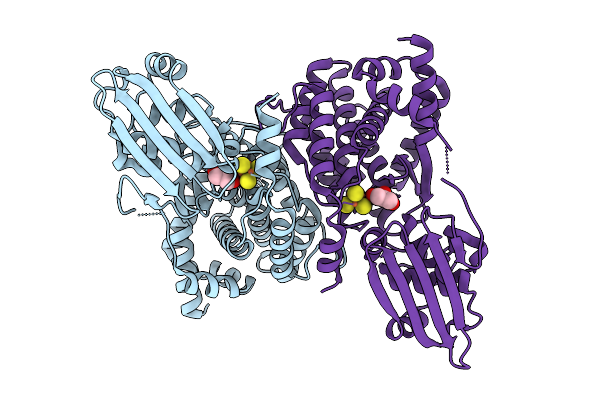
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-11-05 Classification: PROTEIN BINDING Ligands: ZN |
 |
Crystal Structure Of Cyua C289A Mutant From Methanococcus Maripaludis With [4Fe-4S] Clusters In Complex With Glycerol
Organism: Methanococcus maripaludis s2
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LYASE Ligands: SF4, GOL |
 |
Crystal Structure Of Cyua C289A Mutant From Methanococcus Maripaludis With [4Fe-4S] Clusters In Complex With Ethylene Glycol
Organism: Methanococcus maripaludis s2
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: LYASE Ligands: SF4, EDO |
 |
Organism: Rhodococcus rhodochrous
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: BIOSYNTHETIC PROTEIN Ligands: CL, FMN, PUJ, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: ISOMERASE Ligands: A1IU3 |

