Search Count: 283
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: CO, CA, CL, NA |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: NI, CL, CA |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: CU |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: ZN, CA, NA |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: ZN, CL, BS3, NA |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: BS3, NA, CO |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: BS3, NI, CA |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: METAL BINDING PROTEIN Ligands: CL, NA, BS3 |
 |
Organism: Helicobacter pylori g27, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: HYDROLASE |
 |
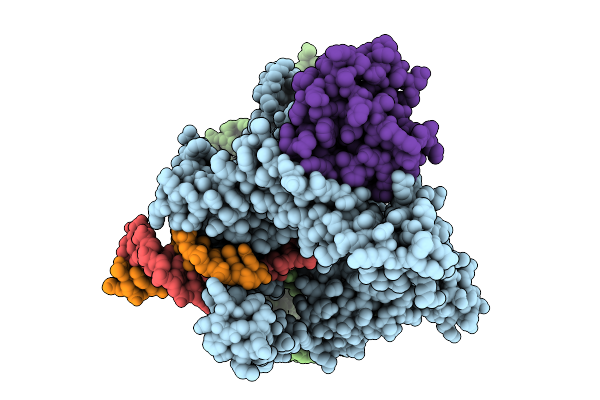
Bacterial Ribosomal 2'-O-Methyltransferase Rsmi In Complex With The Small Ribosomal Subunit
Organism: Escherichia coli, Escherichia coli bw25113
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: AN6, MG |
 |
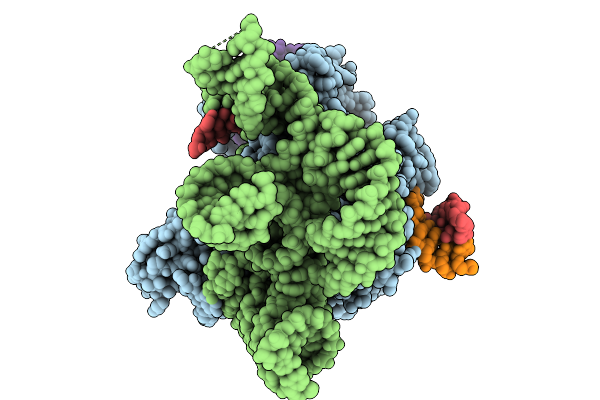
Structure Of Cas12P-Trxa-Guide Rna-Target Dna Complex(29Nt Ts And 11Nt Nts)
Organism: Unidentified, Synthetic construct, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA BINDING PROTEIN/RNA/DNA |
 |
Organism: Unidentified, Synthetic construct, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA BINDING PROTEIN/RNA/DNA |
 |
Crystal Structure Of Escherichia Coli Groel With Magnesium Ions And A Phosphorylated Serine Residue-3.2A
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: CHAPERONE Ligands: HEZ, MG, BME, TAM |
 |
Crystal Structure Of Escherichia Coli Groel With Magnesium Ions And A Phosphorylated Serine Residue-3.0 A
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: CHAPERONE Ligands: BME, HEZ, MG |
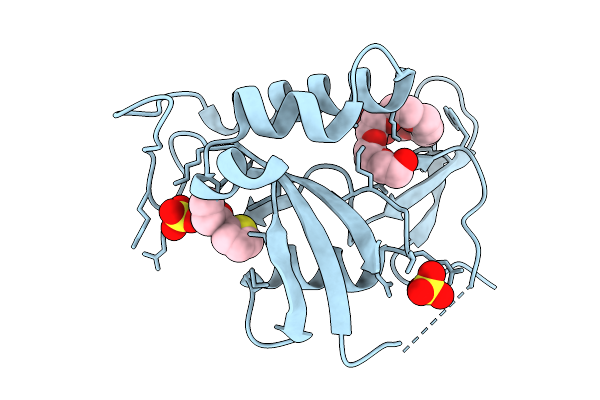 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: ISOMERASE Ligands: QCF, PE8, SO4 |
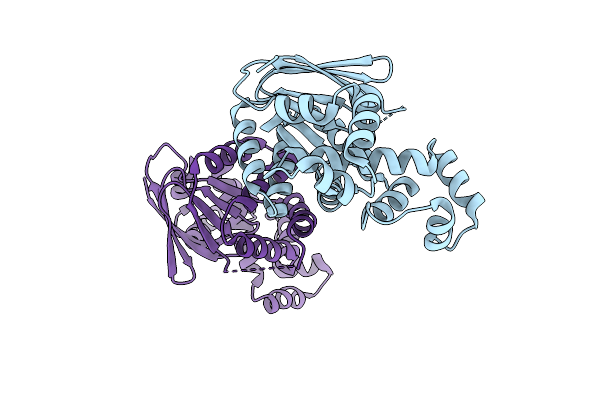 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSCRIPTION |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of E.Coli Ac4C Amidohydrolase Yqfb In Complex With Cytidine
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE Ligands: CTN, MG |
 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: HYDROLASE Ligands: CTN, MG, A1EHT |

