Search Count: 350
All
Selected
 |
Crystal Structure Of Altererythrobacter Sp. S1-5 Tryptophan Halogenase Putative
Organism: Altererythrobacter sp. s1-5
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2026-02-11 Classification: TRANSFERASE Ligands: GOL, CL, ACT |
 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Pentameric Form
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Resolution:2.10 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The K2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The O2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
 |
Organism: Erythrobacter sanguineus
Method: ELECTRON MICROSCOPY Resolution:2.74 Å Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: BCL, A1ELD |
 |
Organism: Erythrobacter sanguineus
Method: ELECTRON MICROSCOPY Resolution:2.27 Å Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: BCL, A1EL2, A1ELD, 8K6, PGV, LMT, BPH, UQ8, FE, H4X, PEF, PO4, LHG |
 |
Organism: Erythrobacter cryptus dsm 12079
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-02-19 Classification: HYDROLASE Ligands: MES |
 |
Crystal Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form At Ph 4.6
Organism: Erythrobacter
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA |
 |
Crystal Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form At Ph 8.8
Organism: Erythrobacter
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA |
 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Pentameric Form At Ph 8.0
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Resolution:2.63 Å Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT |
 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Pentameric Form At Ph 4.3
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2024-04-24 Classification: MEMBRANE PROTEIN Ligands: LMT, LFA |
 |
Crystal Structure Of Light-Activated Dna-Binding Protein El222 From Erythrobacter Litoralis Crystallized And Measured In Dark.
Organism: Erythrobacter litoralis htcc2594
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-07-05 Classification: TRANSCRIPTION Ligands: FMN, CL, MG, MES |
 |
Crystal Structure Of Light-Activated Dna-Binding Protein El222 From Erythrobacter Litoralis Crystallized In Dark, Measured Illuminated.
Organism: Erythrobacter litoralis htcc2594
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-07-05 Classification: TRANSCRIPTION Ligands: FMN, CL, MG, MES |
 |
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2022-02-23 Classification: HYDROLASE Ligands: GOL, EDO, 6NA, DMS, CCN, D8F, SO4, DIO, BUA, NPO, NA |
 |
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-07-14 Classification: HYDROLASE Ligands: NPO, 6NA, PEG, EDO, DMS, DIO, GOL, SO4, PGE, NA, MES |
 |
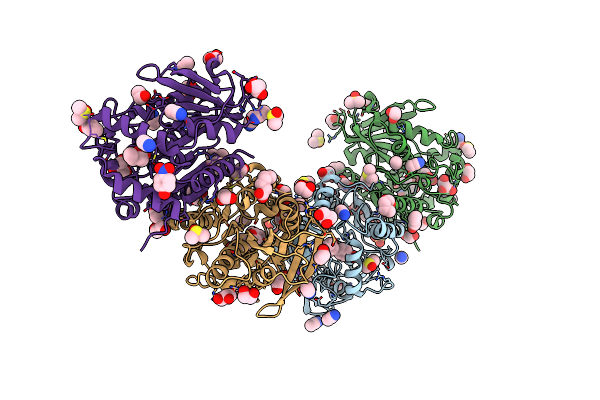
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2021-07-14 Classification: HYDROLASE Ligands: D8F, DMS, GOL, EDO, DIO, NPO, SO4 |
 |
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-07-14 Classification: HYDROLASE Ligands: GOL, EDO, 6NA, DMS, CCN, D8F, SO4, DIO, BUA, NPO, NA |
 |
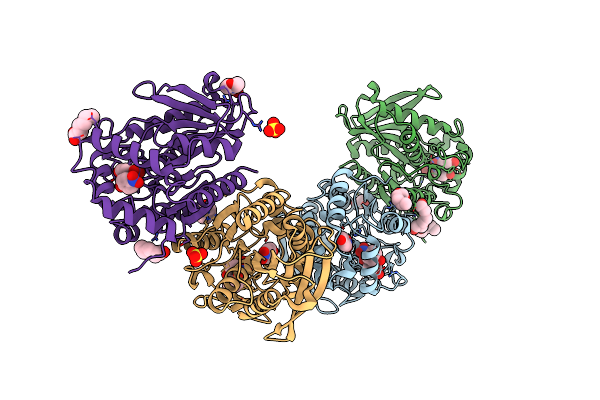
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: D8F, SO4, EDO, GOL, 6NA |
 |
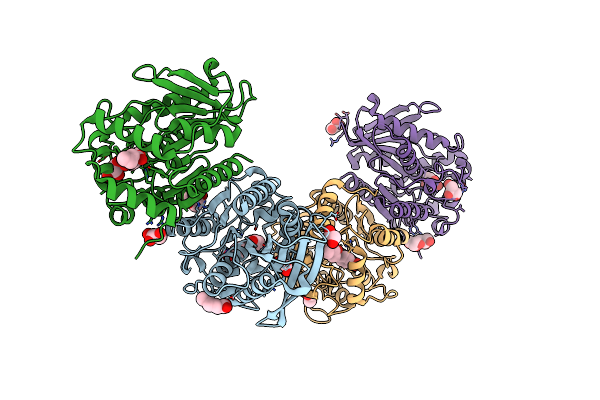
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: NPO, 6NA, PGE, EDO, SO4, MES |
 |
Organism: Erythrobacter longus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2020-07-08 Classification: HYDROLASE Ligands: D8F, 6NA, EDO, GOL, 2FK |

