Search Count: 13
 |
Catalytic S88C Mutant Of Gut Microbial Sulfatase From Bacteroides Fragilis Cag:558
Organism: Bacteroides fragilis cag:558
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-11-11 Classification: HYDROLASE Ligands: CA |
 |
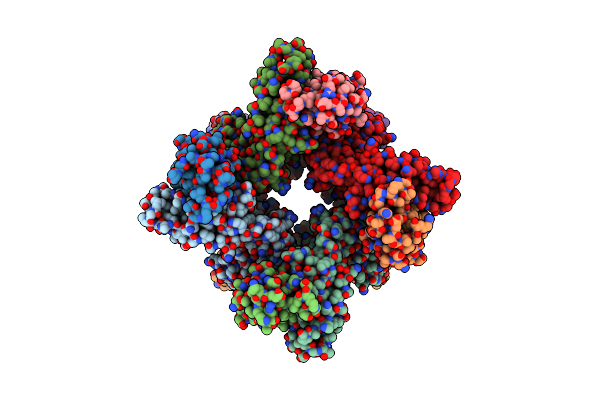
Organism: Hungatella hathewayi
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-11-11 Classification: HYDROLASE Ligands: CA |
 |
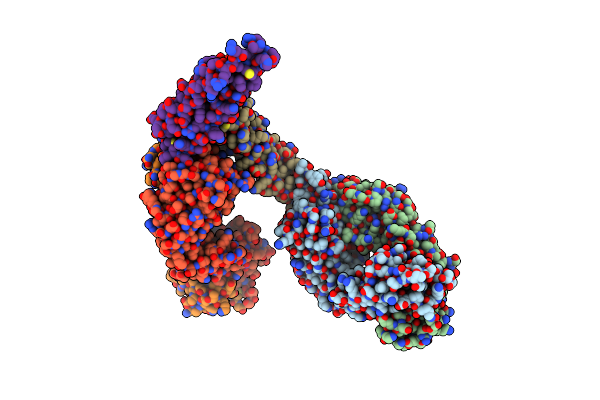
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-04-22 Classification: ANTIVIRAL PROTEIN |
 |
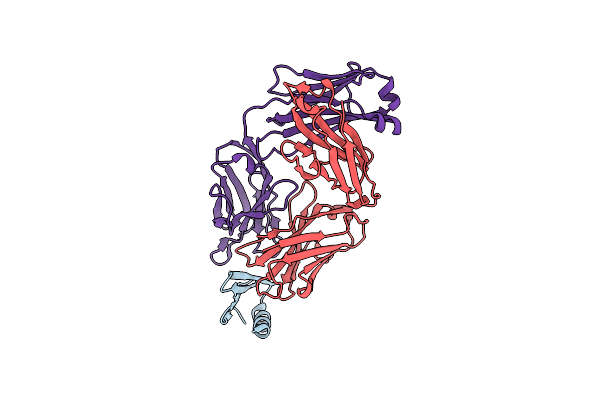
Crystal Structure Of A Computationally Designed Immunogen S2_1.2 In Complex With Its Elicited Antibody C57
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-04-22 Classification: IMMUNE SYSTEM |
 |
De Novo Protein Design Enables The Precise Induction Of Rsv-Neutralizing Antibodies
Organism: Mus musculus, Human respiratory syncytial virus a2
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-04-15 Classification: IMMUNE SYSTEM |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2020-04-15 Classification: DE NOVO PROTEIN |
 |
Organism: Faecalibacterium prausnitzii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-10-30 Classification: HYDROLASE |
 |
Organism: Uncultured clostridium sp.
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-30 Classification: HYDROLASE Ligands: CA |
 |
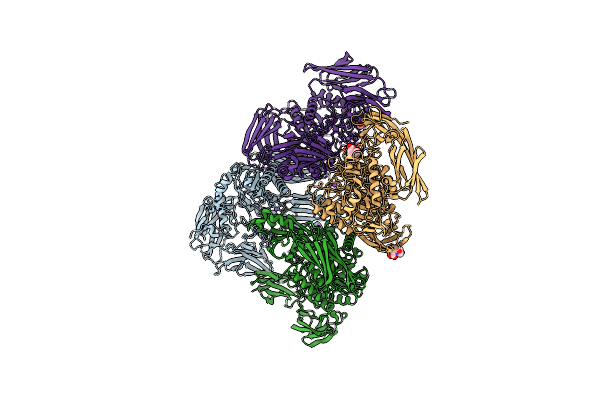
The Crystal Structure Of Parabacteroides Merdae Beta-Glucuronidase (Gus) With Glycerol In Active-Site
Organism: Parabacteroides merdae cl03t12c32
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2019-05-01 Classification: HYDROLASE Ligands: K, GOL, PRO, NA |
 |
Organism: Parabacteroides merdae atcc 43184
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-02-27 Classification: HYDROLASE Ligands: BCN, NA |
 |
Crystal Structure Of Fmn-Binding Beta-Glucuronidase From Facaelibacterium Prausnitzii L2-6
Organism: Faecalibacterium prausnitzii l2-6
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2019-01-30 Classification: HYDROLASE Ligands: FMN |
 |
Crystal Structure Of Fmn-Binding Beta-Glucuronidase From Ruminococcus Gnavus
Organism: [ruminococcus] gnavus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-01-30 Classification: HYDROLASE Ligands: FMN, CA |
 |
Organism: Roseburia hominis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-01-30 Classification: HYDROLASE Ligands: CA, FMN |

