Search Count: 40
 |
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: MG |
 |
Structure Of The Legionella Phosphocholine Hydrolase Lem3 In Complex With Its Substrate Rab1
Organism: Legionella pneumophila, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-04-12 Classification: HYDROLASE Ligands: CA, GDP, OJU |
 |
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-04-12 Classification: METAL BINDING PROTEIN Ligands: MG, PGE, SO4 |
 |
Organism: Legionella pneumophila subsp. pneumophila (strain philadelphia 1 / atcc 33152 / dsm 7513), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-06-10 Classification: SIGNALING PROTEIN Ligands: GDP, MG |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-06-03 Classification: ANTIBIOTIC Ligands: FAD, GOL, PO4, PEG |
 |
Crystal Structure Of P. Aeruginosa Pqsl In Complex With 2-Aminobenzoylacetate
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-06-03 Classification: ANTIBIOTIC Ligands: FAD, 61M, PGE, PO4, MES, PEG |
 |
Crystal Structure Of Pqsl, A Probable Fad-Dependent Monooxygenase From Pseudomonas Aeruginosa - New Refinement
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-04-25 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of Beta-Galactosidase C (Bgac) From Bacillus Circulans Atcc 31382
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-08-27 Classification: HYDROLASE Ligands: PG0 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-05-18 Classification: HYDROLASE Ligands: MN |
 |
Organism: Coryneform bacterium
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-01-12 Classification: HYDROLASE |
 |
Organism: Coryneform bacterium
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2011-01-12 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Coccidioides immitis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2002-12-18 Classification: HYDROLASE |
 |
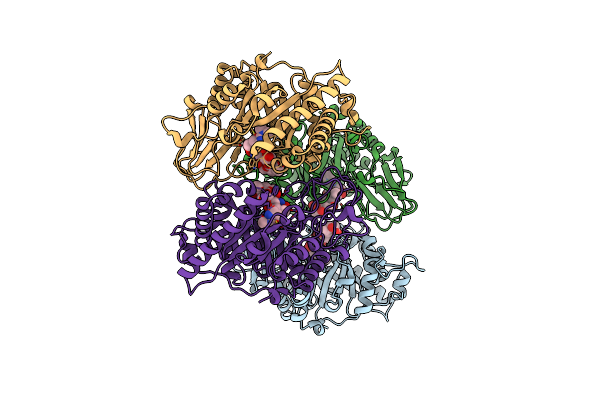
Organism: Coccidioides immitis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-12-18 Classification: HYDROLASE |
 |
Organism: Coccidioides immitis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2002-09-25 Classification: HYDROLASE Ligands: AMI |
 |
Organism: Lactobacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2002-03-13 Classification: LYASE |
 |
Organism: Lactobacillus sp.
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2002-03-13 Classification: LYASE |
 |
Structure Of The D53,54N Mutant Of Histidine Decarboxylase Bound With Histidine Methyl Ester At-170 C
Organism: Lactobacillus sp. 30a
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2002-03-13 Classification: LYASE Ligands: PVH |
 |
Structure Of The D53,54N Mutant Of Histidine Decarboxylase Bound With Histidine Methyl Ester At 25 C
Organism: Lactobacillus sp. 30a
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2002-03-13 Classification: LYASE Ligands: PVH |
 |
Organism: Lactobacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2001-03-21 Classification: LYASE |
 |
Organism: Sambucus ebulus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2001-01-24 Classification: HYDROLASE Ligands: GAL |

