Search Count: 30
 |
Organism: Synthetic construct, Bos taurus
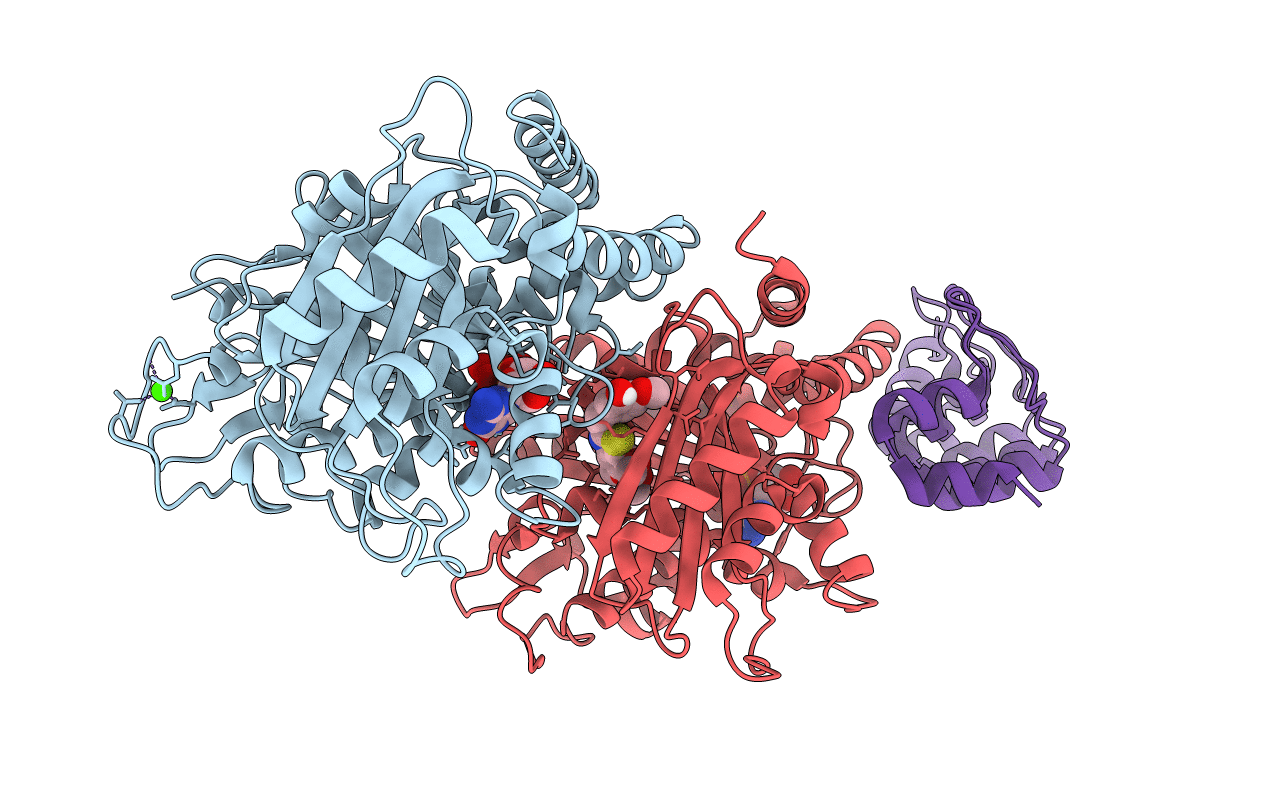
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2022-03-30 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, I8R |
 |
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2022-03-30 Classification: CELL CYCLE Ligands: GTP, MG, GDP, I8N |
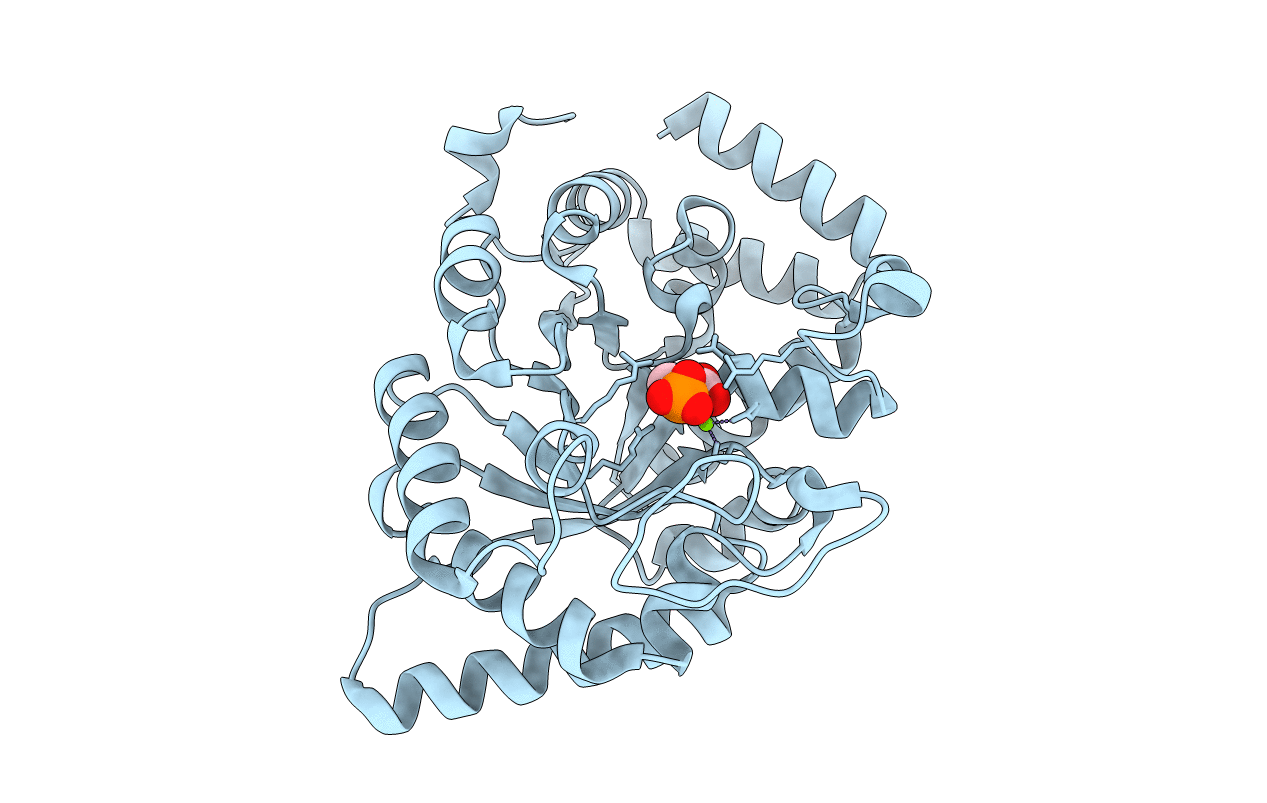 |
Crystal Structure Of The Phosphoenolpyruvate-Binding Domain Of Enzyme I In Complex With Phosphoenolpyruvate From The Thermoanaerobacter Tengcongensis Pep-Sugar Phosphotransferase System (Pts)
Organism: Thermoanaerobacter tengcongensis
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2011-06-15 Classification: TRANSFERASE Ligands: PEP, MG |
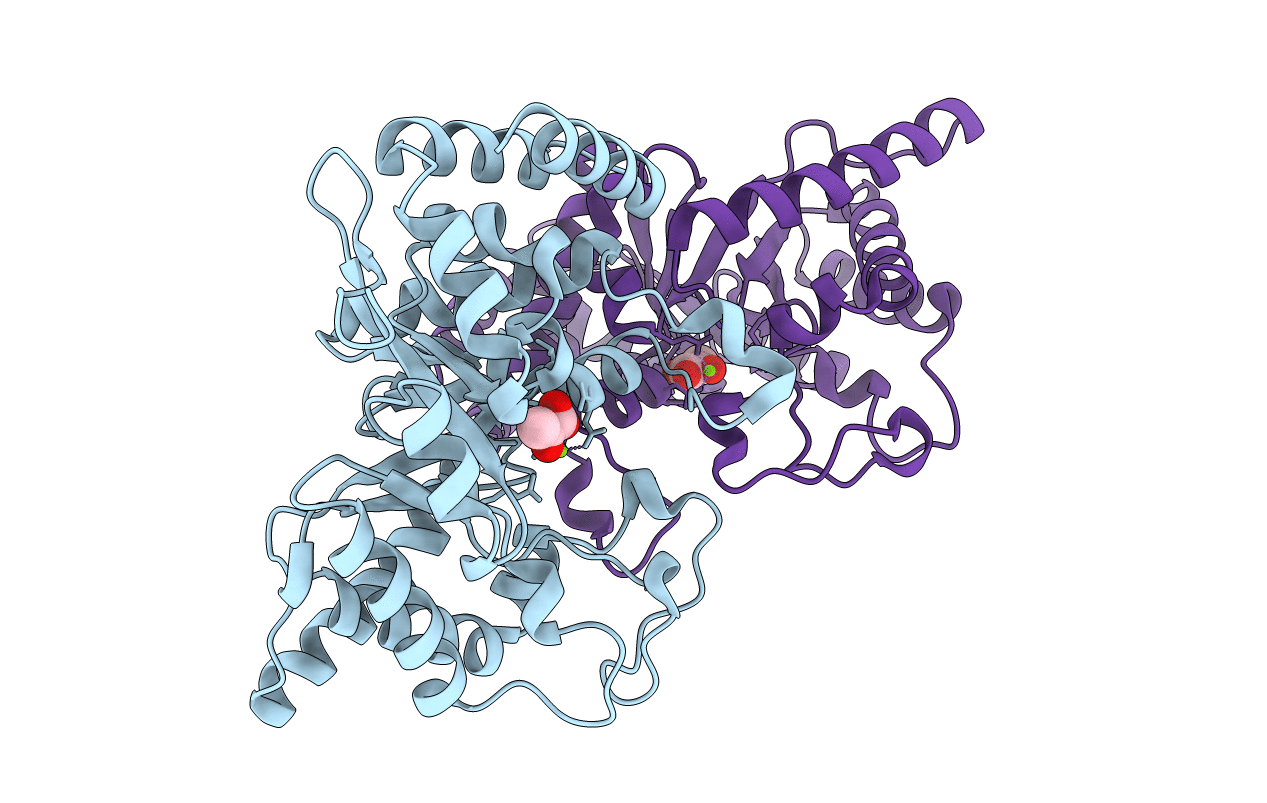 |
Crystal Structure From The Phosphoenolpyruvate-Binding Domain Of Enzyme I In Complex With Pyruvate From The Thermoanaerobacter Tengcongensis Pep-Sugar Phosphotransferase System (Pts)
Organism: Thermoanaerobacter tengcongensis
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2011-06-15 Classification: TRANSFERASE Ligands: MG, PYR |
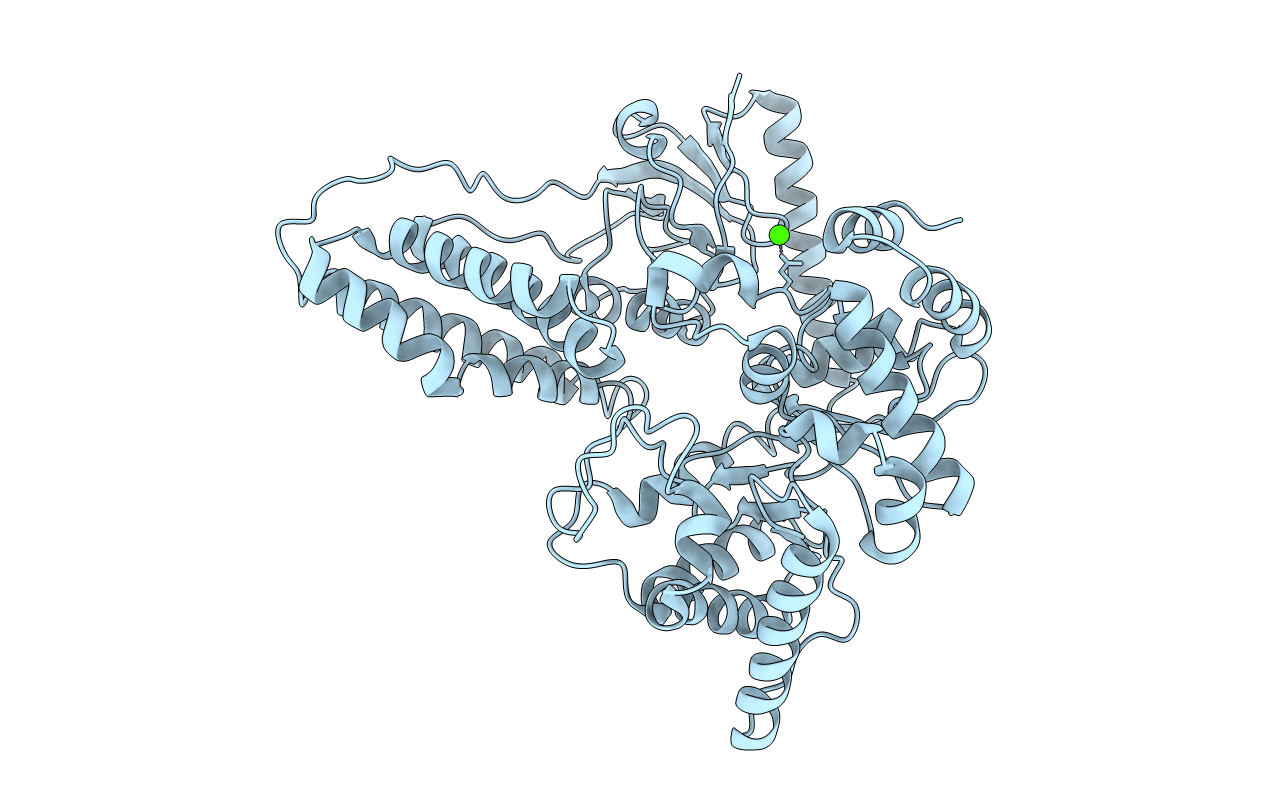 |
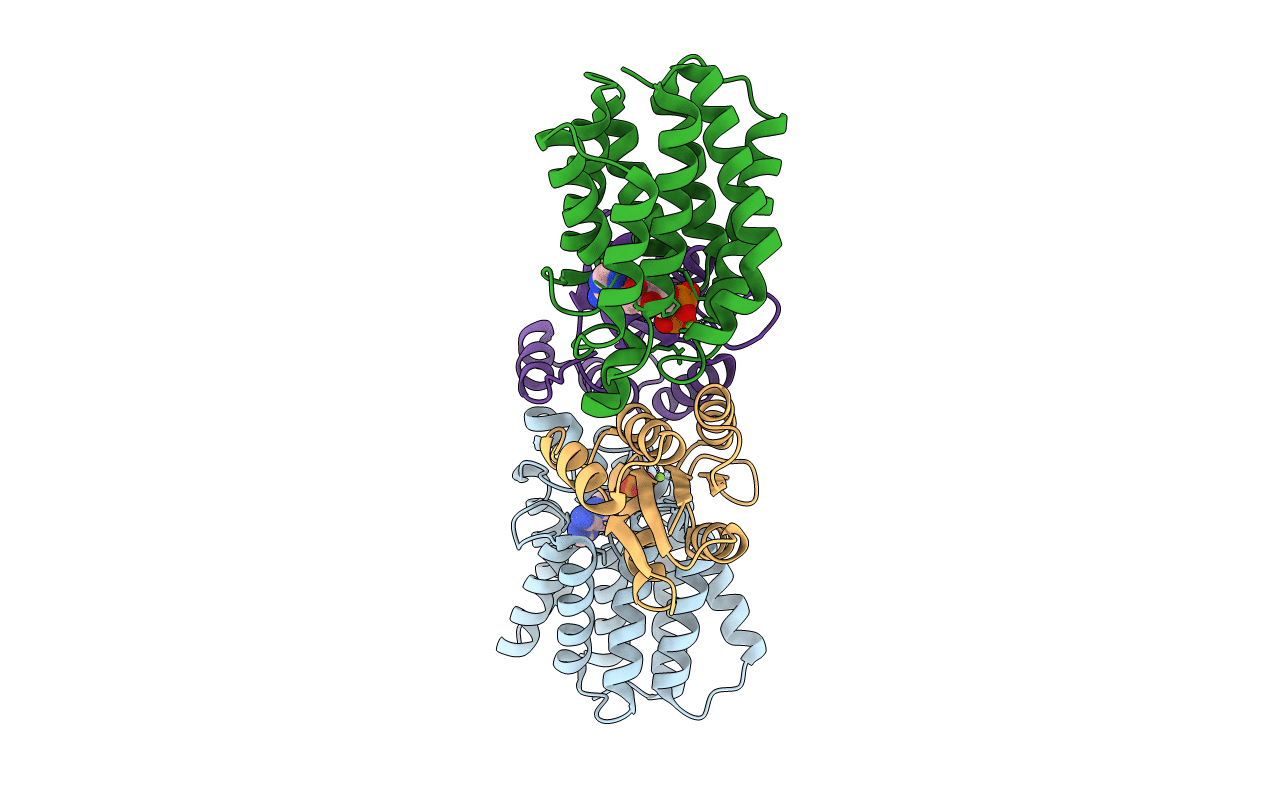
Crystal Structure Of Enzyme I Of The Phosphoenolpyruvate:Sugar Phosphotransferase System In The Dephosphorylated State
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2009-10-13 Classification: TRANSFERASE Ligands: CA |
 |
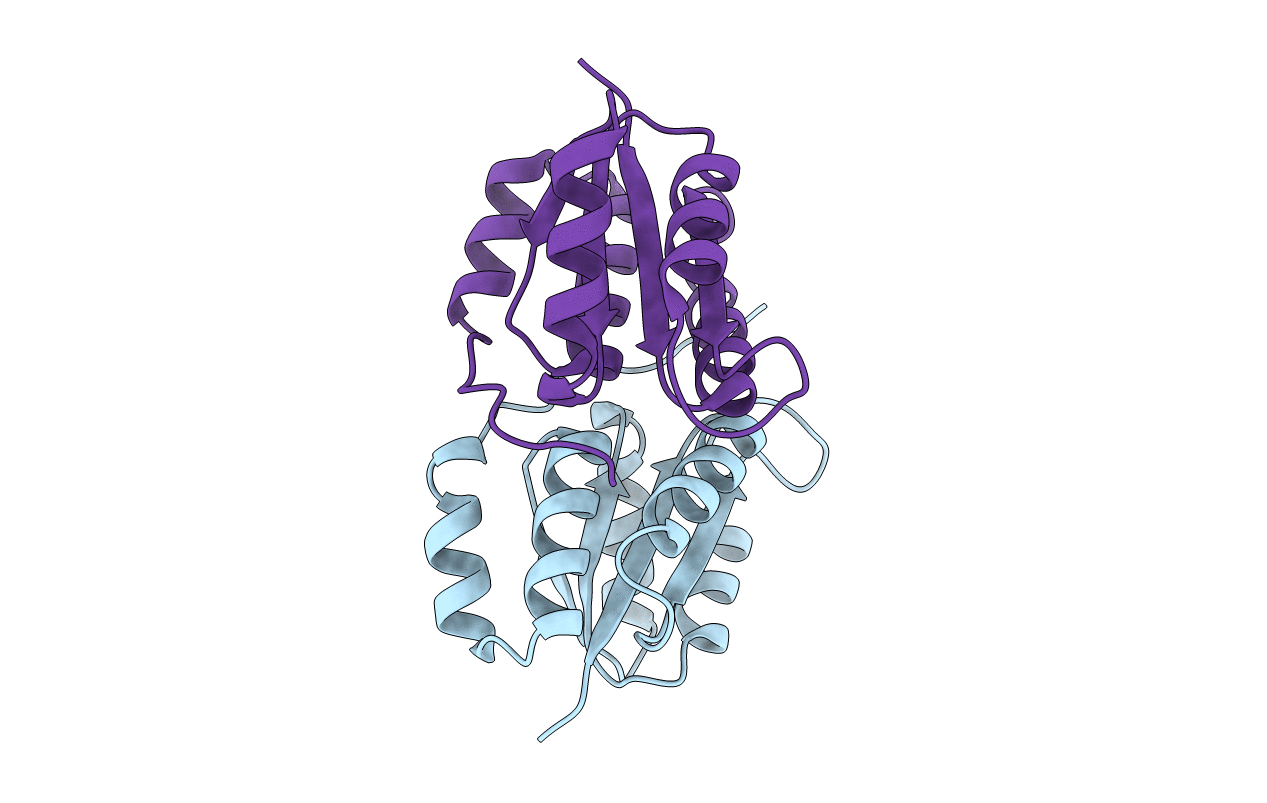
Structure Of A Transient Complex Between Dha-Kinase Subunits Dham And Dhal From Lactococcus Lactis
Organism: Lactococcus lactis subsp. lactis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-10-21 Classification: TRANSFERASE Ligands: MG, ADP |
 |
Organism: Lactococcus lactis subsp. lactis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-10-21 Classification: TRANSFERASE Ligands: 2HA |
 |
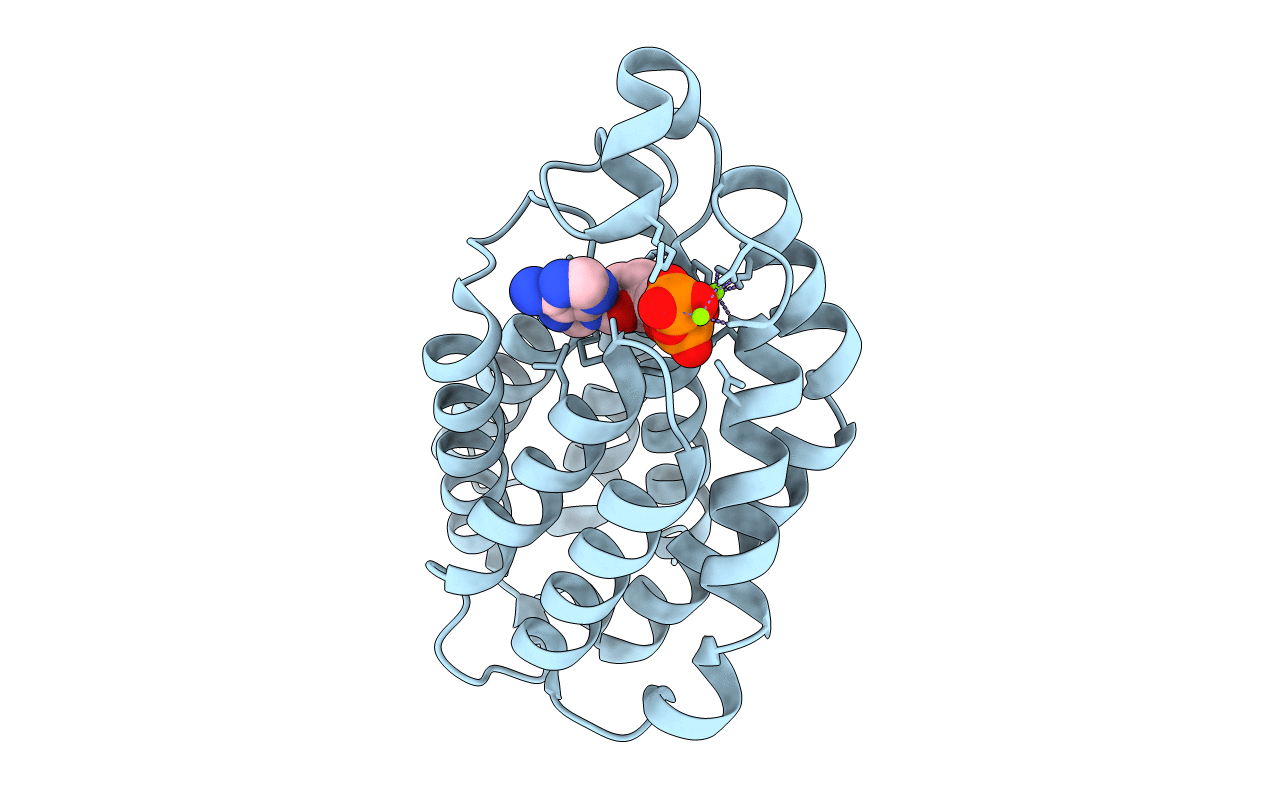
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2008-10-21 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2006-06-22 Classification: TRANSFERASE Ligands: ADP, MG |
 |
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2006-06-13 Classification: TRANSFERASE Ligands: SO4 |
 |
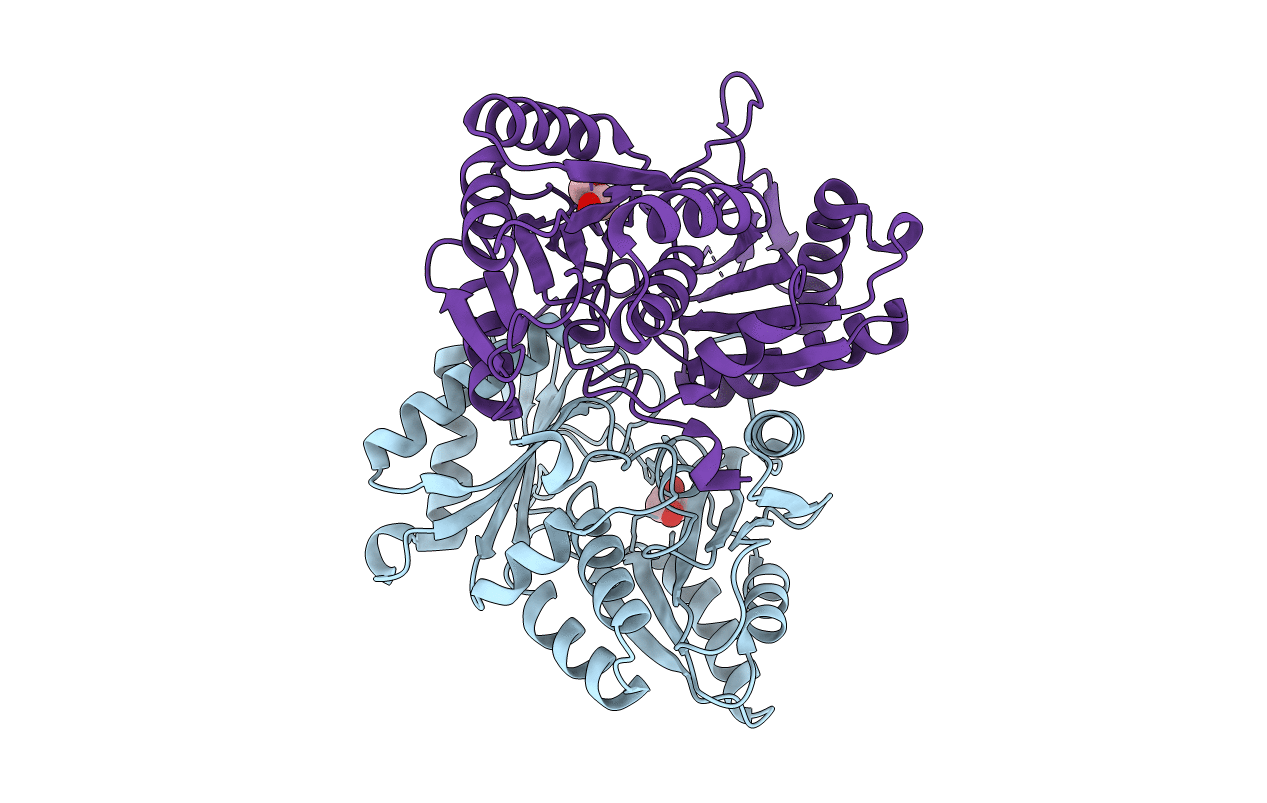
Organism: Lactococcus lactis subsp. lactis il1403
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-06-13 Classification: TRANSCRIPTION |
 |
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-06-13 Classification: TRANSFERASE Ligands: GOL |
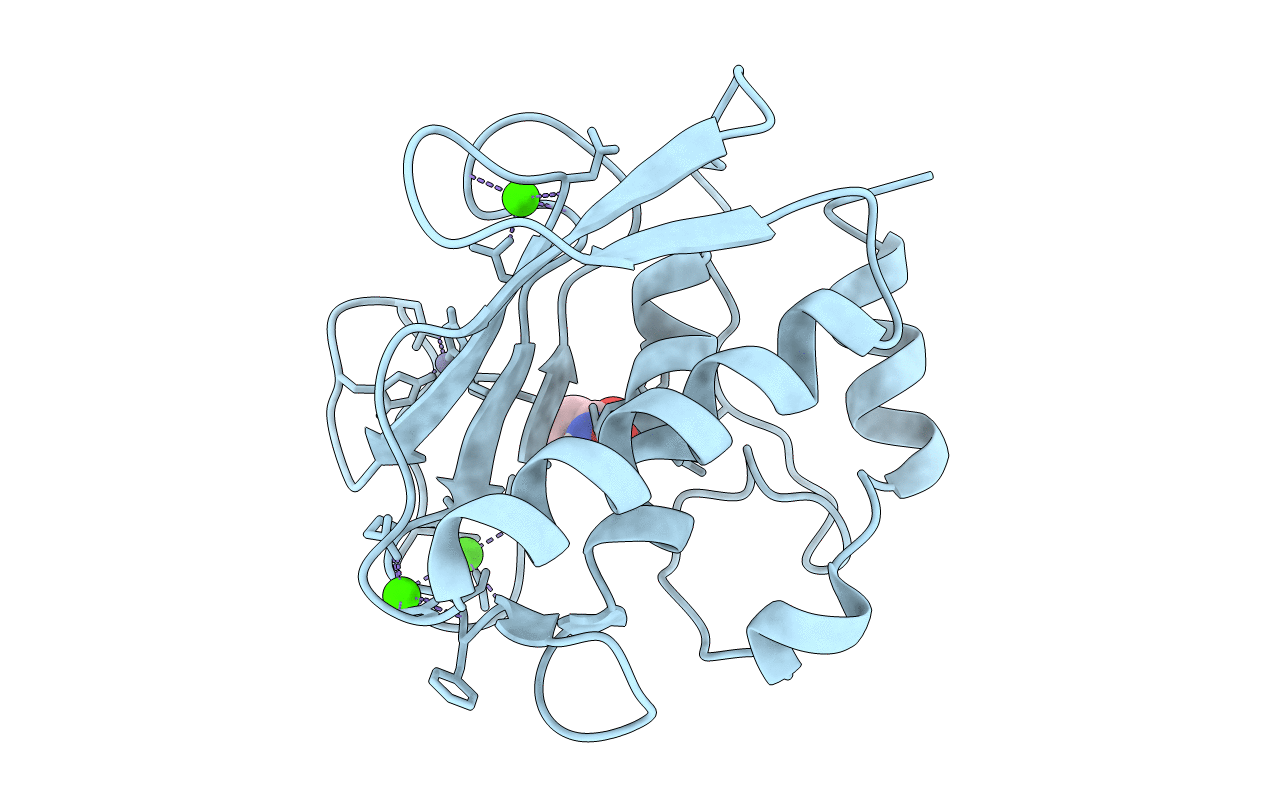 |
Crystal Structure Of The Catalytic Domain Of Human Mmp12 Complexed With Acetohydroxamic Acid At Atomic Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2005-04-26 Classification: HYDROLASE Ligands: ZN, CA, HAE |
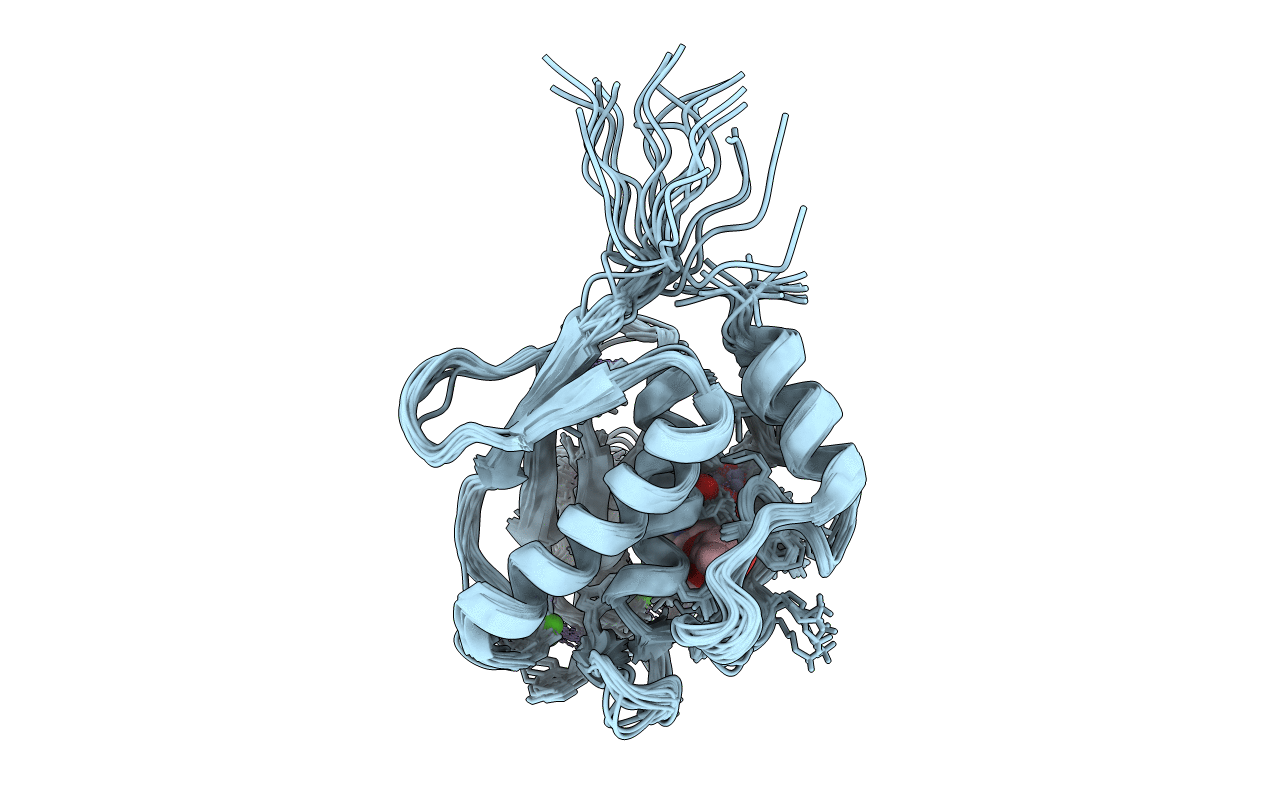 |
Solution Structure Of Matrix Metalloproteinase 12 (Mmp12) In The Presence Of N-Isobutyl-N-[4-Methoxyphenylsulfonyl]Glycyl Hydroxamic Acid (Nngh)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2005-04-19 Classification: HYDROLASE Ligands: ZN, CA, NGH |
 |
Solution Structure Of Mmp12 In The Presence Of N-Isobutyl-N-4-Methoxyphenylsulfonyl]Glycyl Hydroxamic Acid (Nngh)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2005-04-19 Classification: HYDROLASE Ligands: ZN, CA, NGH |
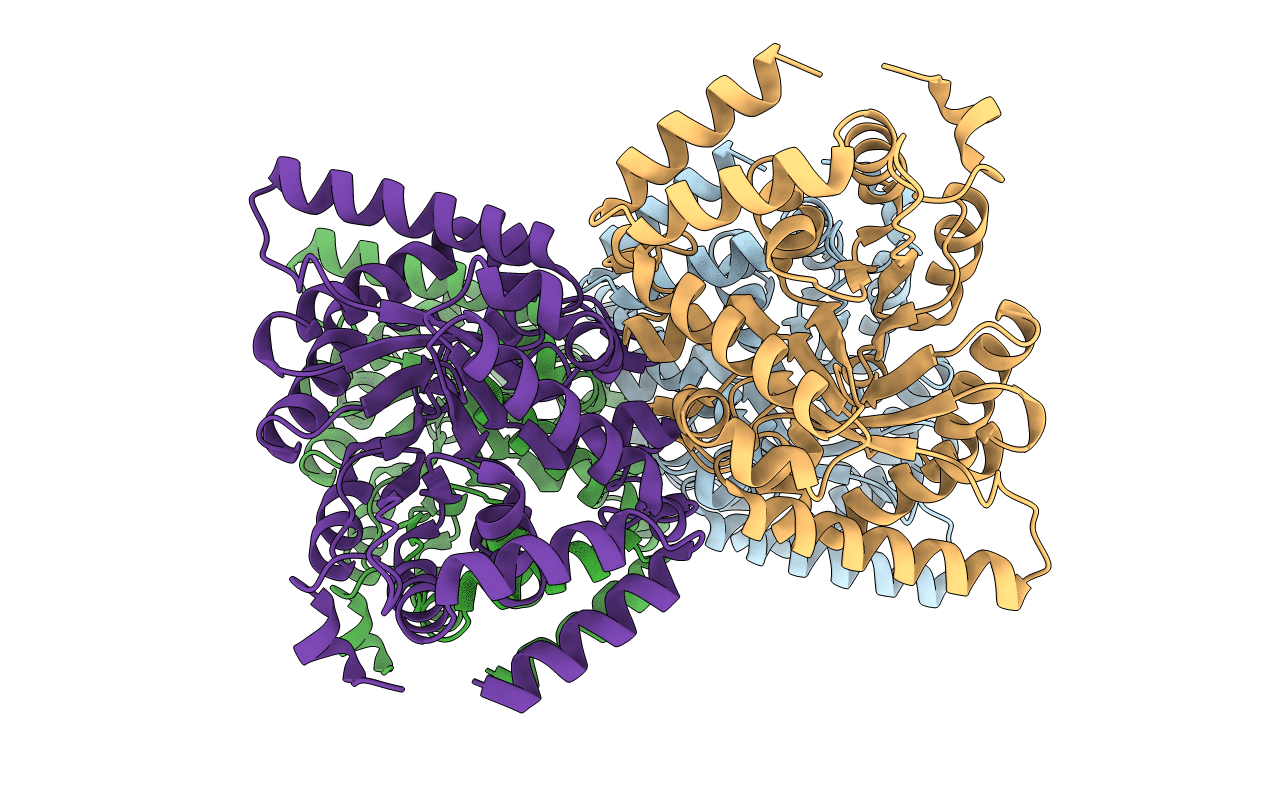 |
Crystal Structure Of The Phosphoenolpyruvate-Binding Enzyme I-Domain From The Thermoanaerobacter Tengcongensis Pep: Sugar Phosphotransferase System (Pts)
Organism: Thermoanaerobacter tengcongensis
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2005-02-02 Classification: TRANSFERASE |
 |
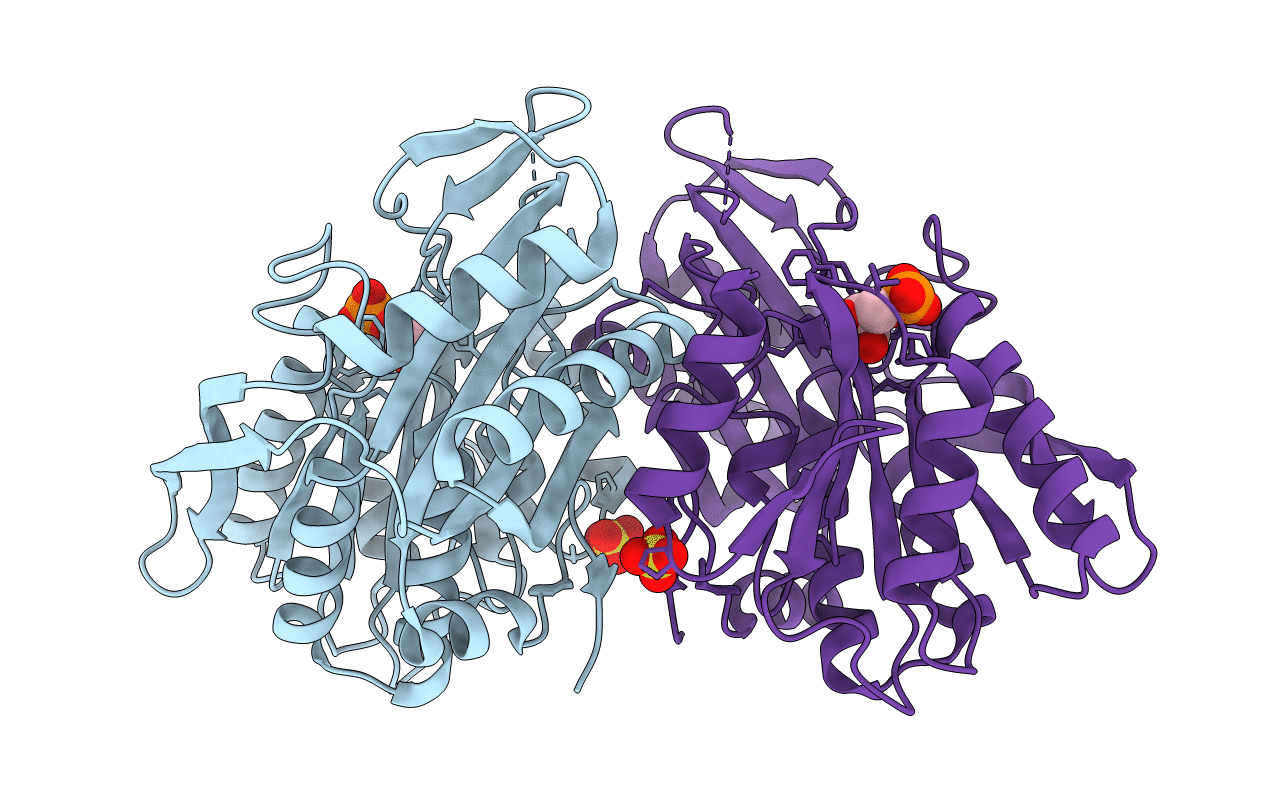
Crystal Structure Of The Catalytic Domain Of Human Mmp12 Complexed With The Inhibitor Nngh At 1.3 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2004-12-14 Classification: HYDROLASE Ligands: ZN, CA, NGH |
 |
Crystal Structure Of The Dihydroxyacetone Kinase From E. Coli In Complex With Dihydroxyacetone-Phosphate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-09-24 Classification: TRANSFERASE Ligands: G3H, SO4 |
 |
Crystal Structure Of The Dihydroxyacetone Kinase From E. Coli In Complex With Glyceraldehyde
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-09-24 Classification: TRANSFERASE Ligands: GOL, SO4 |
 |
Crystal Structure Of The Catalytic Domain Of Human Matrix Metalloproteinase 10
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-04-06 Classification: HYDROLASE Ligands: ZN, CA, NGH |

