Search Count: 7
 |
Organism: Vaccinia virus western reserve
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-05-18 Classification: HYDROLASE Ligands: SO4, GOL |
 |
The Crystal Structure Of Pf-8, The Dna Polymerase Accessory Subunit From Kaposi S Sarcoma-Associated Herpesvirus
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2010-05-12 Classification: REPLICATION |
 |
The Crystal Structure Of Pf-8, The Dna Polymerase Accessory Subunit From Kaposi'S Sarcoma-Associated Herpesvirus
Organism: Human herpesvirus 8
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2009-11-24 Classification: REPLICATION |
 |
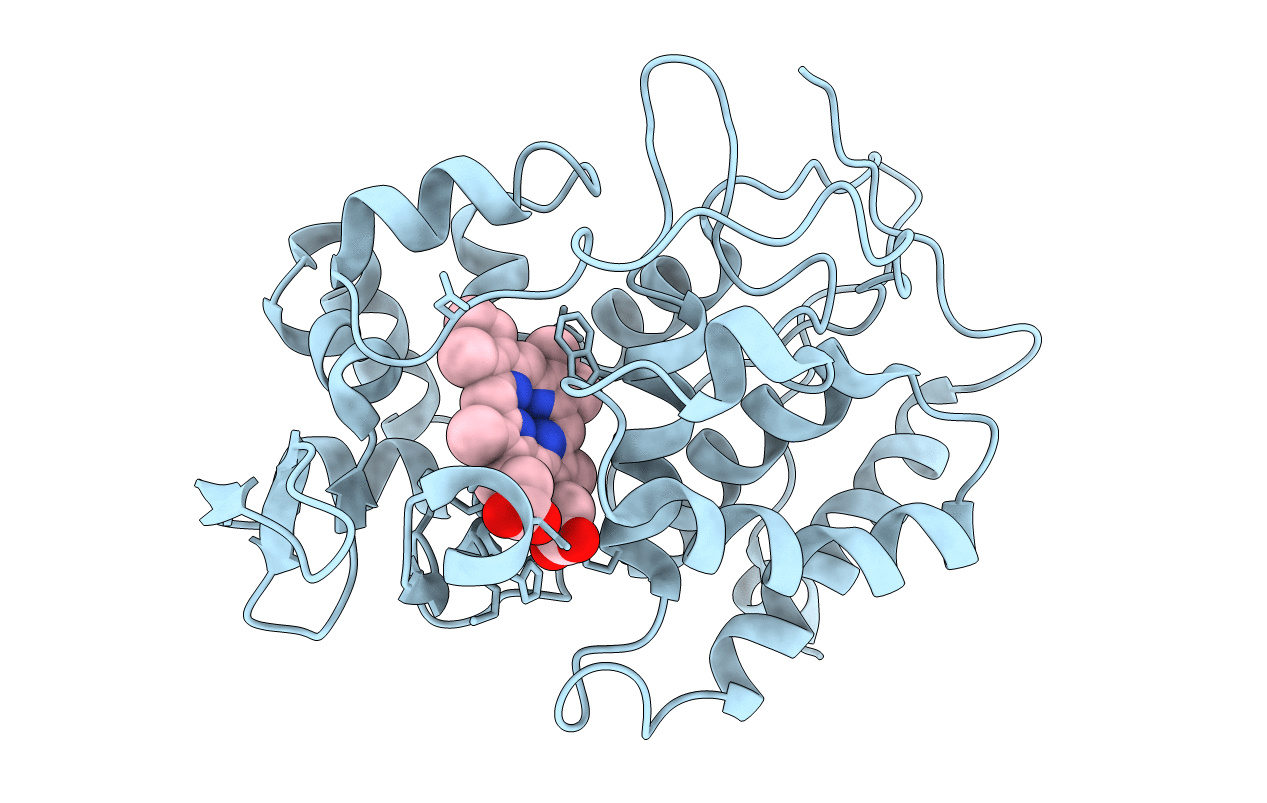
Laue Diffraction Study On The Structure Of Cytochrome C Peroxidase Compound I
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2001-07-26 Classification: OXIDOREDUCTASE Ligands: HEM, O |
 |
Histidine 52 Is A Critical Residue For Rapid Formation Of Cytochrome C Peroxidase Compound I
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1993-10-31 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
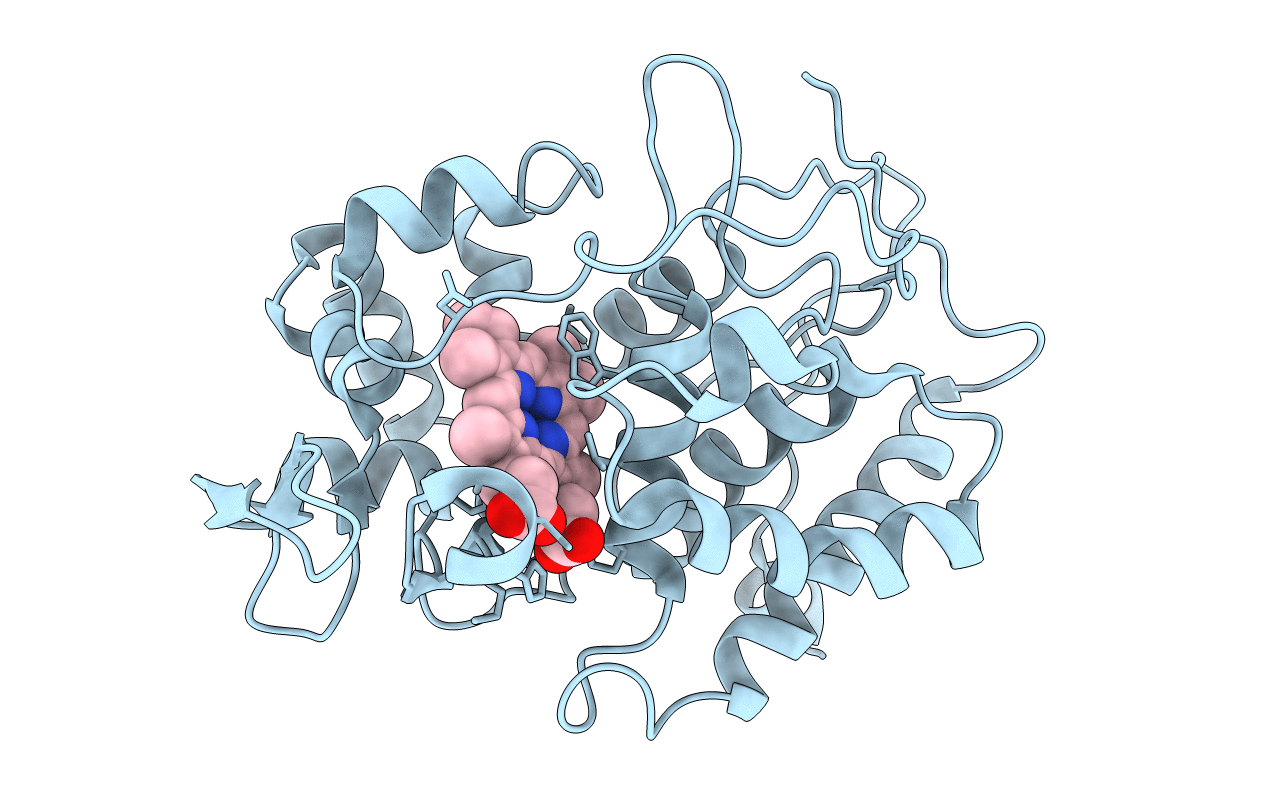
Effect Of Arginine-48 Replacement On The Reaction Between Cytochrome C Peroxidase And Hydrogen Peroxide
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1993-10-31 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Effect Of Arginine-48 Replacement On The Reaction Between Cytochrome C Peroxidase And Hydrogen Peroxide
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1993-10-31 Classification: OXIDOREDUCTASE Ligands: HEM |

