Search Count: 13
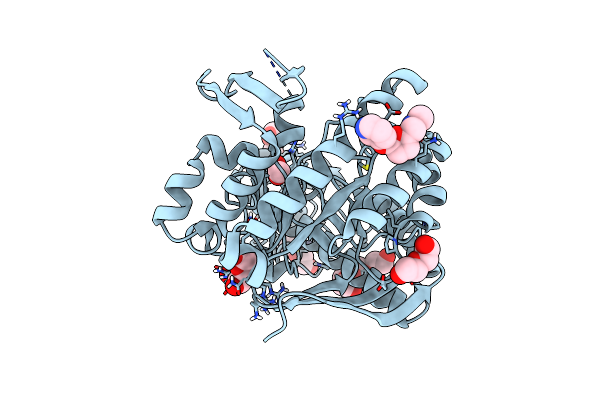 |
Crystal Structure Of Tgt In Complex With Methyl({[5-(Pyridin-3-Yloxy)Furan-2-Yl]Methyl})Amine
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-03-20 Classification: TRANSFERASE Ligands: ZN, PEG, DMS, F63, PG4, ACT, PGE |
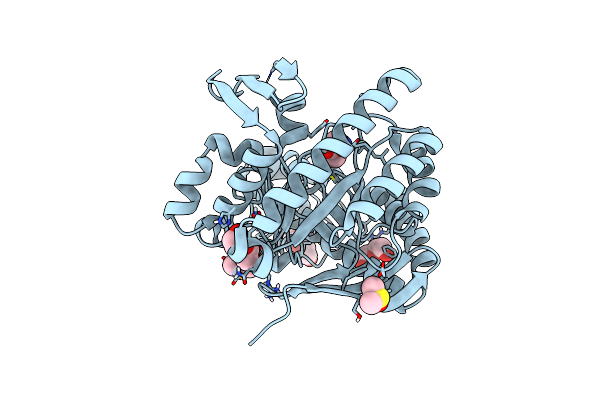 |
Crystal Structure Of Tgt In Complex With 4-(Aminomethane)Cyclohexane-1-Carboxylic Acid
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2018-05-16 Classification: TRANSFERASE Ligands: ZN, AMH, PEG, DMS, PGE |
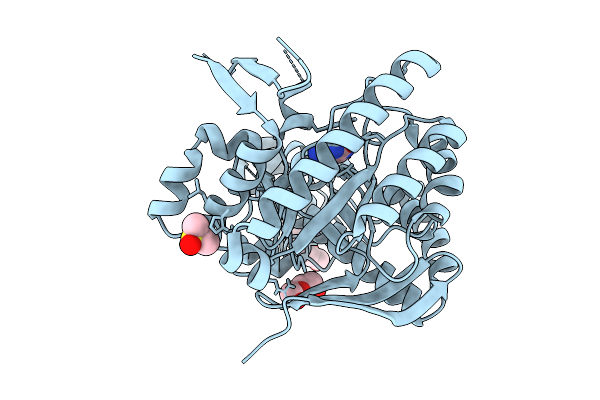 |
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2018-03-07 Classification: TRANSFERASE Ligands: ZN, DMS, PEG, GUN, PGE |
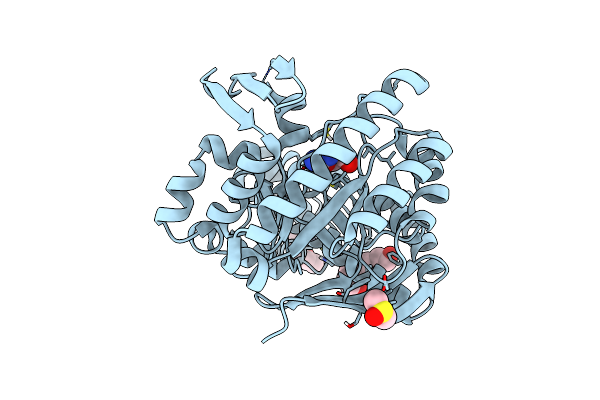 |
Organism: Zymomonas mobilis subsp. mobilis zm4 = atcc 31821
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2018-03-07 Classification: TRANSFERASE Ligands: ZN, DMS, GGB, PGE, PG4 |
 |
Crystal Structure Of Tgt In Complex With 2,6-Dioxy-8-Azapurine, 2,6-Dioxy-8-Azapurine, 2,6-Dioxy-8-Azapurine
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-03-07 Classification: TRANSFERASE Ligands: ZN, PEG, DMS, AZA |
 |
Crystal Structure Of Tgt In Complex With 3-Pyridinecarboxylic Acid, 6-(Dimethylamino)
Organism: Zymomonas mobilis subsp. mobilis (strain atcc 31821 / zm4 / cp4)
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2017-07-19 Classification: TRANSFERASE Ligands: ZN, DMS, 46L, PEG, PG4 |
 |
Identification Of Small Molecule Inhibitors Selective For Apo(A) Kringles Kiv-7, Kiv-10 And Kv.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-07-16 Classification: HYDROLASE Ligands: CL, BU6 |
 |
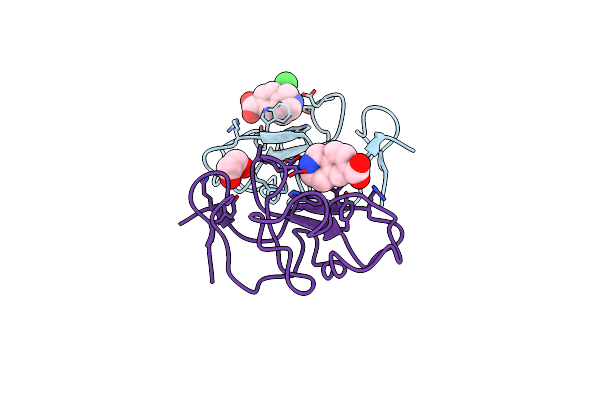
Identification Of Small Molecule Inhibitors Selective For Apo(A) Kringles Kiv-7, Kiv-10 And Kv.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-07-16 Classification: HYDROLASE Ligands: CPF, SO4 |
 |
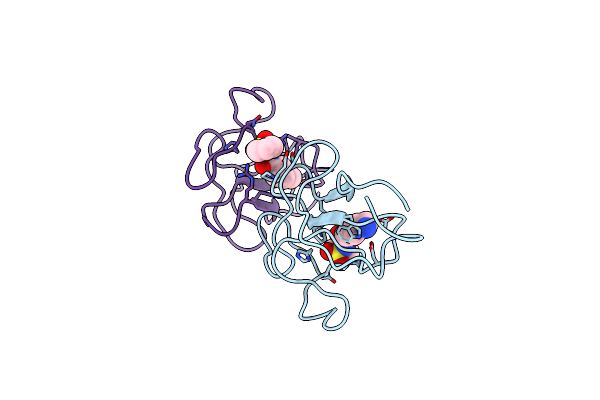
Identification Of Small Molecule Inhibitors Selective For Apo(A) Kringles Kiv-7, Kiv-10 And Kv.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-07-16 Classification: HYDROLASE Ligands: CL, HKY, EDO |
 |
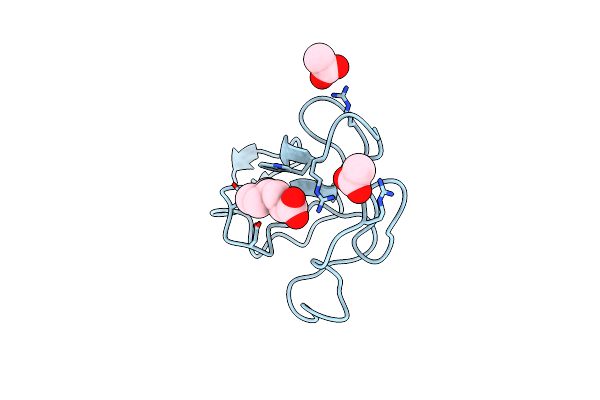
Identification Of Small Molecule Inhibitors Selective For Apo(A) Kringles Kiv-7, Kiv-10 And Kv.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-07-09 Classification: HYDROLASE Ligands: 5C3 |
 |
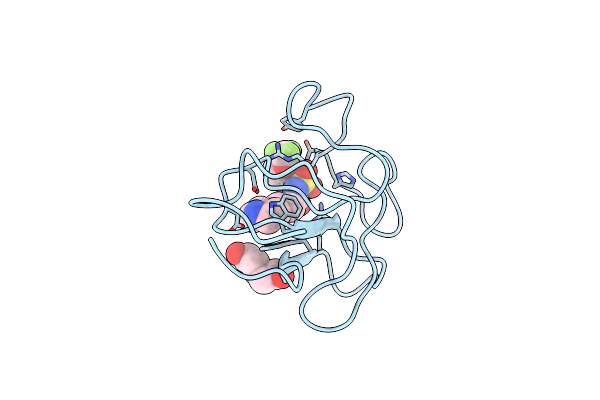
Identification Of Small Molecule Inhibitors Selective For Apo(A) Kringles Kiv-7, Kiv-10 And Kv.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-07-09 Classification: HYDROLASE Ligands: ACT, BV7 |
 |
Identification Of Small Molecule Inhibitors Selective For Apo(A) Kringles Kiv-7, Kiv-10 And Kv.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-07-09 Classification: HYDROLASE Ligands: CL, BV5, PEG |
 |

