Search Count: 14
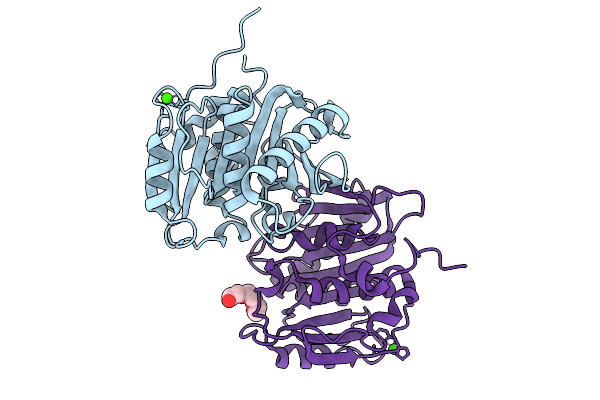 |
Organism: Actinoplanes sp. dh11
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: CA, PGE, CL |
 |
Crystal Structure Of An Alpha/Beta-Hydrolase Enzyme From Chloroflexus Sp. Ms-G (202)
Organism: Chloroflexus sp. ms-g
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-12-28 Classification: HYDROLASE |
 |
Crystal Structure Of An Alpha/Beta-Hydrolase Enzyme From Candidatus Kryptobacter Tengchongensis (306)
Organism: Candidatus kryptobacter tengchongensis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PO4 |
 |
Crystal Structure Of A Cutinase Enzyme From Marinactinospora Thermotolerans Dsm45154 (606)
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2022-12-28 Classification: HYDROLASE |
 |
Organism: Saccharopolyspora flava
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PG4 |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PEG |
 |
Organism: Thermobifida fusca
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: PG4 |
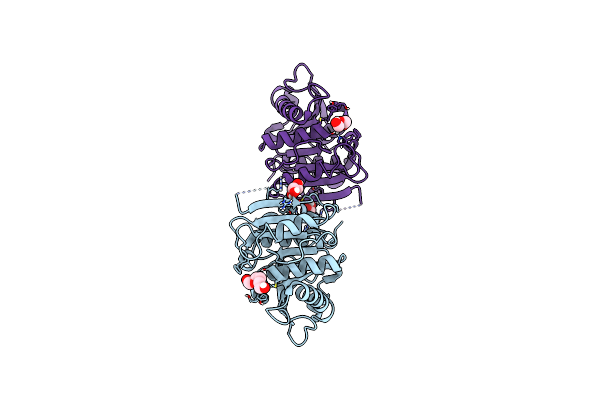 |
Organism: Thermobifida fusca yx
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: GOL, PG4, PEG |
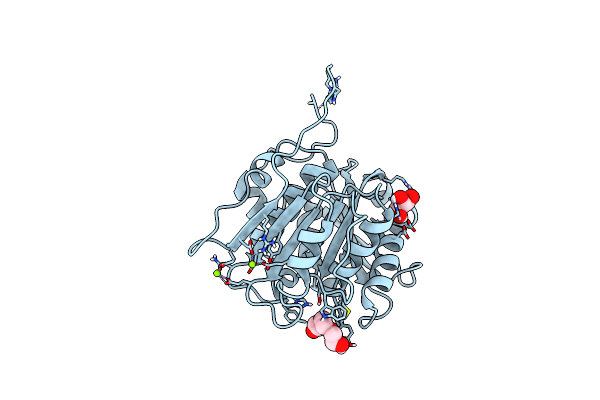 |
Crystal Structure Of A Cutinase Enzyme From Thermobifida Cellulosilytica Tb100 (711)
Organism: Thermobifida cellulosilytica tb100
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: GOL, PG4, MG |
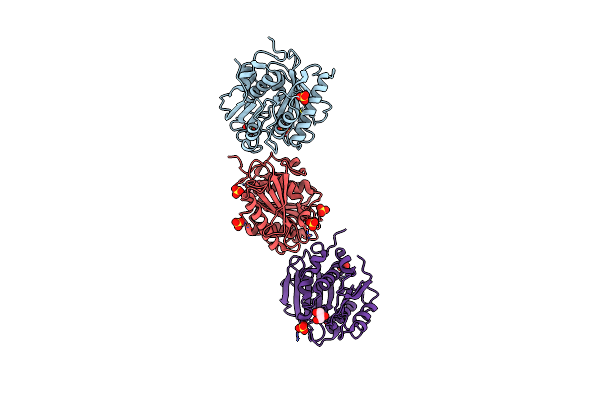 |
Crystal Structure Of A Double Mutant Petase (S238F/W159H) From Ideonella Sakaiensis
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-10-13 Classification: HYDROLASE Ligands: SO4, CL, GOL |
 |
Organism: Ideonella sakaiensis (strain nbrc 110686 / tistr 2288 / 201-f6)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: BEZ, CA |
 |
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: CA |
 |
Organism: Ideonella sakaiensis (strain 201-f6)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-09-30 Classification: STRUCTURAL GENOMICS Ligands: BEZ, CA |
 |
Organism: Ideonella sakaiensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: CA, SO4 |

