Search Count: 21
 |
The Crystal Structure Of A Thermus Thermophilus Trnagly Acceptor Stem Microhelix At 1.6 Angstroem Resolution
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2011-08-10 Classification: RNA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-02-16 Classification: DNA Ligands: NCO, MG, CAC |
 |
Crystal Structure Of The E.Coli Trnaarg Aminoacyl Stem Issoacceptor Rr-1660 At 2.0 Angstroem Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-11-17 Classification: RNA Ligands: GOL |
 |
The 1.2 Angstroem Crystal Structure Of An E.Coli Trnaser Acceptor Stem Microhelix Reveals Two Magnesium Binding Sites
Method: X-RAY DIFFRACTION
Resolution:1.20 Å Release Date: 2009-07-28 Classification: RNA Ligands: MG |
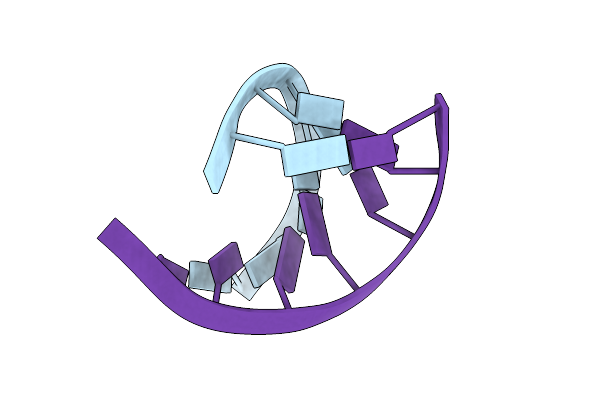 |
Crystal Structure Of A Human Trnagly Acceptor Stem Microhelix (Derived From The Gene Sequence Dg9990) At 1.18 Angstroem Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2009-03-10 Classification: RNA |
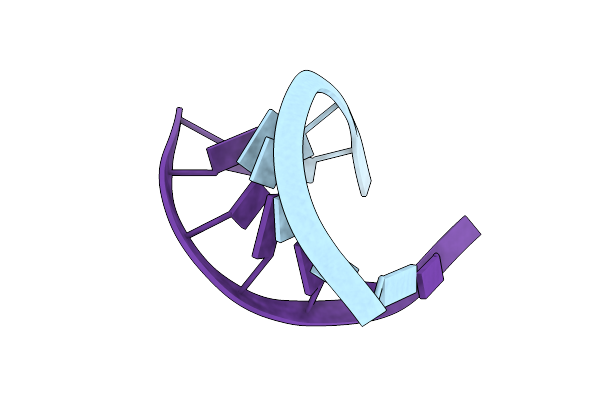 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2008-03-18 Classification: RNA |
 |
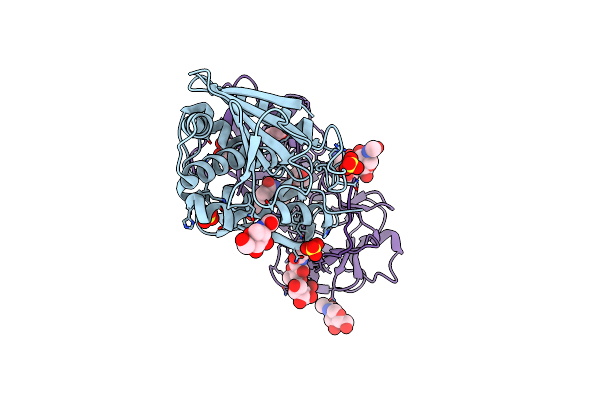
Trnaser Acceptor Stem: Conformation And Hydration Of A Microhelix In A Crystal Structure At 1.8 Angstrom Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-11-06 Classification: RNA |
 |
Organism: Viscum album
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2007-10-30 Classification: HYDROLASE Ligands: NAG, SO4, GOL, CL, SGI |
 |
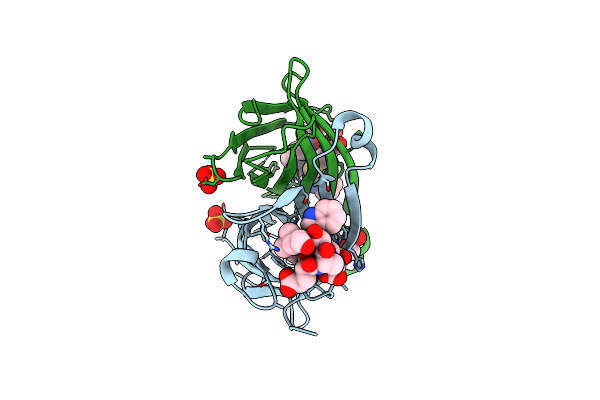
Crystal Structure Of An Escherichia Coli Trnagly Microhelix At 2.0 Angstrom Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-16 Classification: RNA Ligands: MG |
 |
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2007-02-06 Classification: PEPTIDE BINDING PROTEIN Ligands: SO4, GOL |
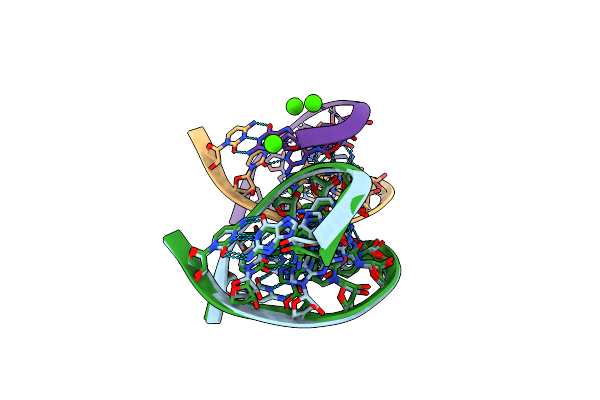 |
Method: X-RAY DIFFRACTION
Resolution:1.40 Å Release Date: 2006-06-27 Classification: RNA Ligands: CA |
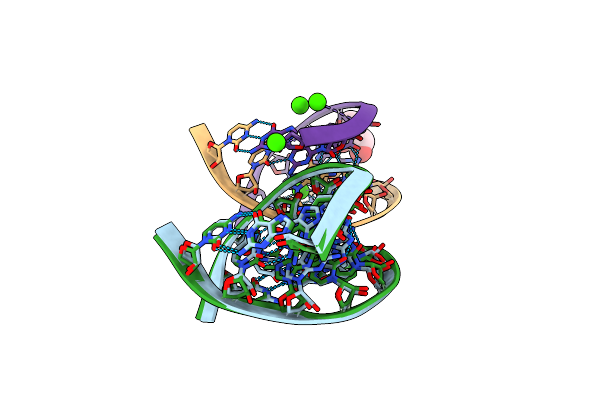 |
Method: X-RAY DIFFRACTION
Resolution:1.35 Å Release Date: 2006-06-27 Classification: RNA Ligands: CA, GOL |
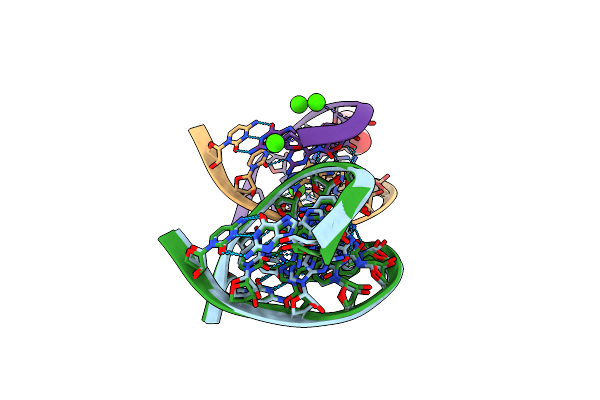 |
Method: X-RAY DIFFRACTION
Resolution:1.40 Å Release Date: 2006-06-27 Classification: RNA Ligands: CA, GOL |
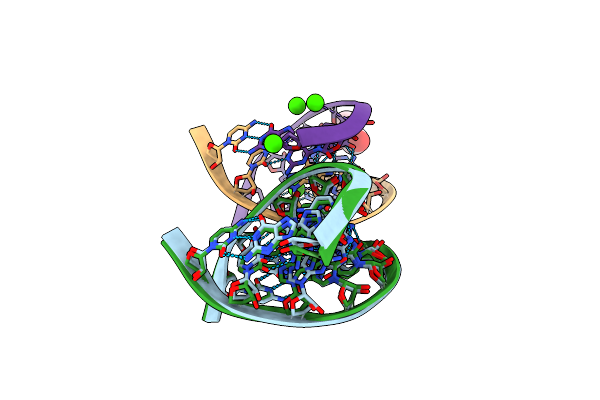 |
Method: X-RAY DIFFRACTION
Resolution:1.30 Å Release Date: 2006-06-27 Classification: RNA Ligands: CA, GOL |
 |
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2006-06-27 Classification: RNA Ligands: CA, GOL |
 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2006-05-30 Classification: RNA Ligands: CA, GOL |
 |
Crystal Structure Of The First Rna Duplex In L-Conformation At 1.9A Resolution
|
 |
Method: X-RAY DIFFRACTION
Resolution:1.60 Å Release Date: 2001-09-14 Classification: RNA Ligands: BA |
 |
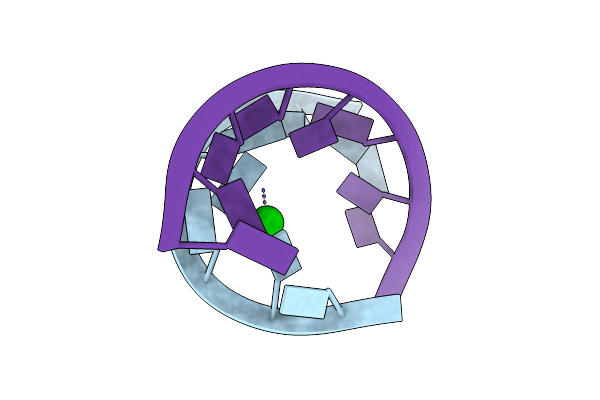
Crystal Structure Of Domain E Of Thermus Flavus 5S Rrna: A Helical Rna-Structure Including A Tetraloop
|
 |
Crystal Structure Of Domain A Of Thermus Flavus 5S Rrna And The Contribution Of Water Molecules To Its Structure
|

