Search Count: 32
 |
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Crystal Structure Of Gh139 Glycoside Hydrolase From Verrucomicrobium Sp. In The Hexagonal Space Group P6522
Organism: Verrucomicrobium sp.
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE, NA, CL |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium (Unmodelled Additional Ligand Density At Active Site)
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE |
 |
Crystal Structure Of Class Ii Sfp Aldolase From Yersinia Aldovae (Yasqia-Zn-So4) With Bound Sulfate Ions
Organism: Yersinia aldovae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-10-25 Classification: LYASE Ligands: SO4, ZN, NA, BTB |
 |
Crystal Structure Of Metal-Dependent Classii Sulfofructosephosphate Aldolase (Sfpa) From Hafnia Paralvei Hpsqia-Zn
Organism: Hafnia paralvei
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-10-25 Classification: LYASE Ligands: ZN |
 |
Crystal Structure Of Metal-Dependent Class Ii Sulfofructose Phosphate Aldolase From Yersinia Aldovae In Complex With Sulfofructose Phosphate (Yasqia-Zn-Sfp)
Organism: Yersinia aldovae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-10-25 Classification: LYASE |
 |
Crystal Structure Of Metal-Dependent Class Ii Sulfofructosephosphate Aldolase From Hafnia Paralvei Hpsqia-Zn In Complex With Dihydroxyacetone Phosphate (Dhap)
Organism: Hafnia paralvei
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-10-25 Classification: LYASE Ligands: ZN, 13P |
 |
Crystal Structure Of The Sulfoquinovosyl Binding Protein Smof Complexed With Sulfoquinovosyl Diacylglycerol
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-04-13 Classification: SUGAR BINDING PROTEIN Ligands: CQI, GLY |
 |
Crystal Structure Of The Sulfoquinovosyl Binding Protein Smof Complexed With Sulfoquinovose
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-04-13 Classification: SUGAR BINDING PROTEIN Ligands: YZT |
 |
Crystal Structure Of The Sulfoquinovosyl Binding Protein Smof Complexed With Sqme
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2022-04-13 Classification: SUGAR BINDING PROTEIN Ligands: DO7 |
 |
Crystal Structure Of Fmnh2-Dependent Monooxygenase From Agrobacterium Tumefaciens For Oxidative Desulfurization Of Sulfoquinovose
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-02-02 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Ligand Free Open Conformation Of Sulfoquinovosyl Binding Protein (Sqbp) From Agrobacterium Tumefaciens
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2022-01-19 Classification: SUGAR BINDING PROTEIN Ligands: ACT |
 |
Crystal Structure Of A Gh31 Family Sulfoquinovosidase Mutant D455N From Agrobacterium Tumefaciens In Complex With Sulfoquinovosyl Glycerol (Sqgro)
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-01-19 Classification: HYDROLASE Ligands: VCW |
 |
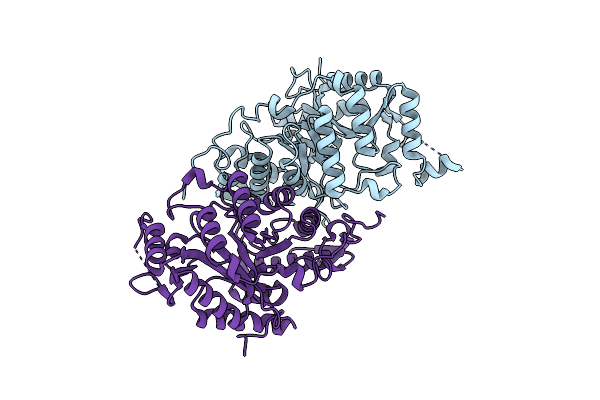
Crystal Structure Of Sq Binding Protein From Agrobacterium Tumefaciens In Complex With Sulfoquinovosyl Glycerol (Sqgro)
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-01-19 Classification: SUGAR BINDING PROTEIN Ligands: VCW, EDO |
 |
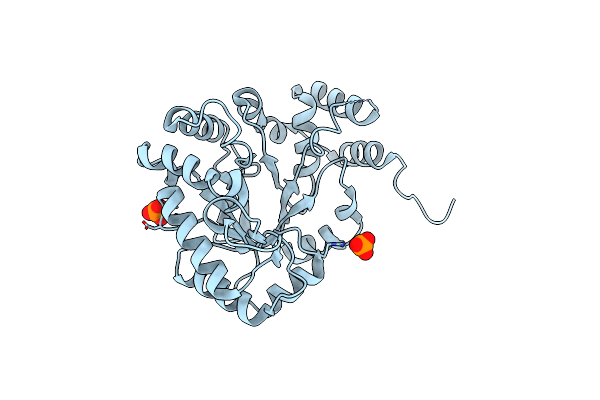
Crystal Structure Of Fmnh2-Dependent Monooxygenase For Oxidative Desulfurization Of Sulfoquinovose
Organism: Rhizobium oryzae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-01-19 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Aldo-Keto Reductase With C-Terminal His Tag From Agrobacterium Tumefaciens
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
Crystal Structure Of Apo Aldo-Keto Reductase From Agrobacterium Tumefaciens
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Aldo-Keto Reductase From Agrobacterium Tumefaciens In A Binary Complex With Nadph
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: PO4 |
 |
Crystal Structure Of Aldo-Keto Reductase From Agrobacterium Tumefaciens In A Ternary Complex With Nadph And Glucose
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: NAP, GLC |

