Search Count: 24
 |
Crystal Structure Of The Alkyltransferase Ribozyme Samuri Co-Crystalized With Prosedma
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-02-05 Classification: RNA Ligands: A1IY6, MG |
 |
Crystal Structure Of The Alkyltransferase Ribozyme Samuri Co-Crystallized With Sam
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-01-22 Classification: RNA Ligands: MG, SAH |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-20 Classification: RNA Ligands: BA |
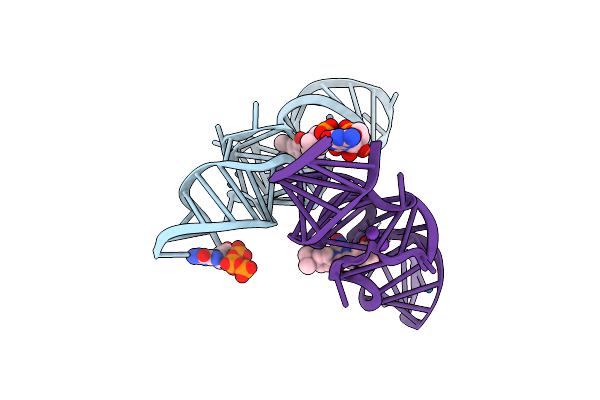 |
Crystal Structure Of Chili Rna Aptamer In Complex With Dmhbo+ (Iridium Hexammine Co-Crystallized Form)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-06-16 Classification: RNA Ligands: V5Z, GTP, K, IR, MG |
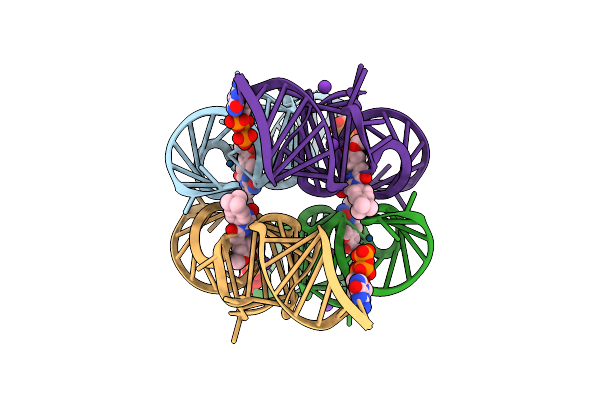 |
Crystal Structure Of Chili Rna Aptamer In Complex With Dmhbo+ (Iridium Iii Hexammine Soaking Crystal Form)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2021-06-16 Classification: RNA Ligands: V5Z, GTP, K, CL, IR |
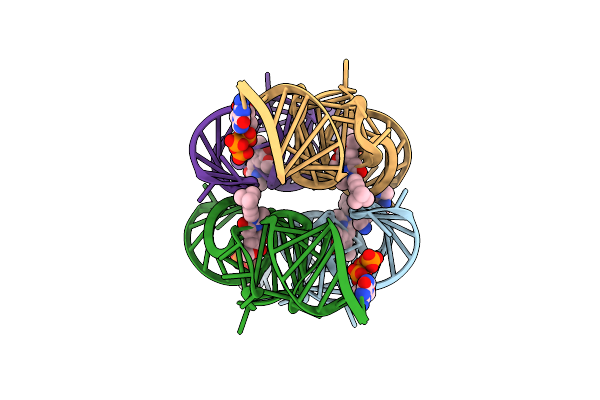 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2021-06-16 Classification: RNA Ligands: GTP, K, V6T, MG, SPM, CL |
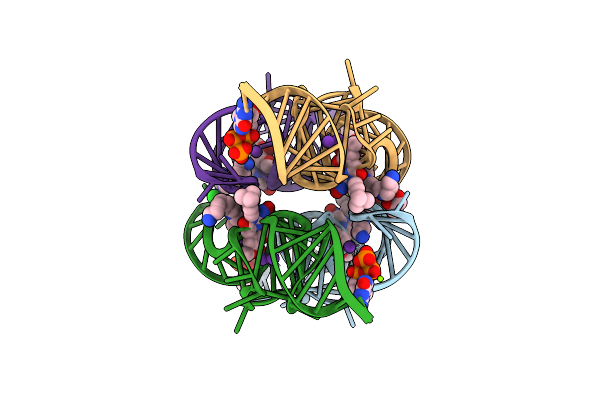 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2021-06-16 Classification: RNA Ligands: GTP, V5Z, SPM, GOL, CL, K, MG |
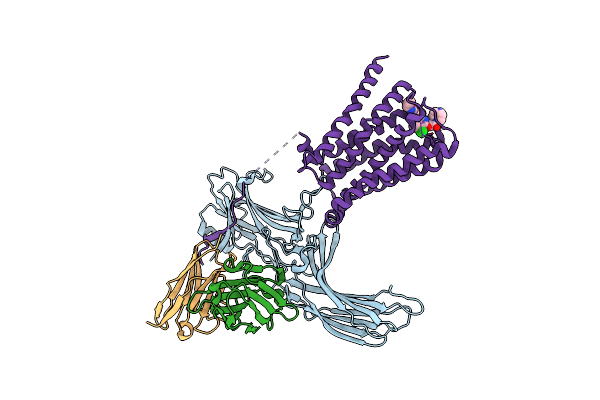 |
Organism: Homo sapiens, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2020-02-26 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: 2CU |
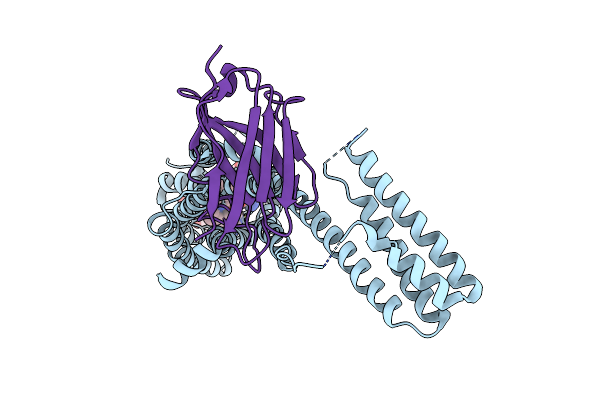 |
Structure Of Synthetic Nanobody-Stabilized Angiotensin Ii Type 1 Receptor Bound To Angiotensin Ii
Organism: Homo sapiens, Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-02-19 Classification: MEMBRANE PROTEIN Ligands: NAG, CL |
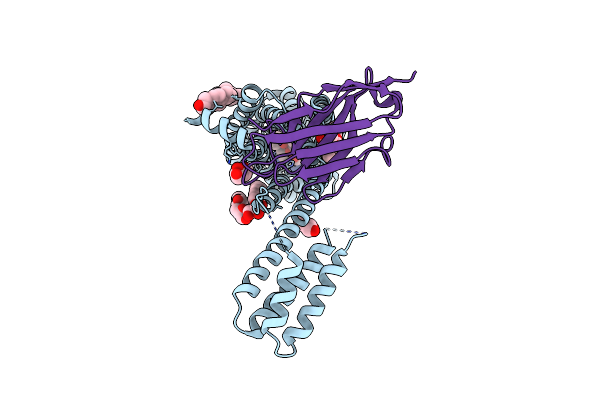 |
Structure Of Synthetic Nanobody-Stabilized Angiotensin Ii Type 1 Receptor Bound To Trv023
Organism: Homo sapiens, Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2020-02-19 Classification: MEMBRANE PROTEIN Ligands: OLC, CLR |
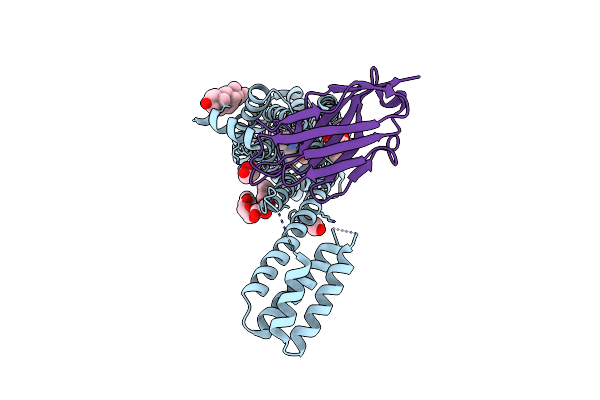 |
Structure Of Synthetic Nanobody-Stabilized Angiotensin Ii Type 1 Receptor Bound To Trv026
Organism: Homo sapiens, Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-02-19 Classification: MEMBRANE PROTEIN Ligands: NAG, OLC, CLR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-08-21 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: DB8 |
 |
Crystal Structure Of The Central Flexible Region Of Ash1 Mrna E3-Localization Element
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-01-18 Classification: RNA Ligands: SO4 |
 |
Crystal Structure Of The Nuclear Complex With She2P And The Ash1 Mrna E3-Localization Element
Organism: Saccharomyces cerevisiae, Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2017-01-18 Classification: TRANSPORT PROTEIN Ligands: EDO, MG, ACT |
 |
Crystal Structure Of The Cytoplasmic Complex With She2P, She3P, And The Ash1 Mrna E3-Localization Element
Organism: Saccharomyces cerevisiae (strain rm11-1a), Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-01-18 Classification: RNA BINDING PROTEIN Ligands: MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-02-15 Classification: SURFACTANT PROTEIN |
 |
Self-Assembly Of Spider Silk Proteins Is Controlled By A Ph-Sensitive Relay
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-05-12 Classification: STRUCTURAL PROTEIN Ligands: EDO, PEG |
 |
Self-Assembly Of Spider Silk Proteins Is Controlled By A Ph-Sensitive Relay
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-05-12 Classification: STRUCTURAL PROTEIN Ligands: PGE |
 |
Self-Assembly Of Spider Silk Proteins Is Controlled By A Ph-Sensitive Relay
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-05-12 Classification: STRUCTURAL PROTEIN Ligands: EDO, PEG, PGE |
 |
Self-Assembly Of Spider Silk Proteins Is Controlled By A Ph-Sensitive Relay
Organism: Euprosthenops australis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2010-05-12 Classification: STRUCTURAL PROTEIN Ligands: EDO, PGE |

