Search Count: 18
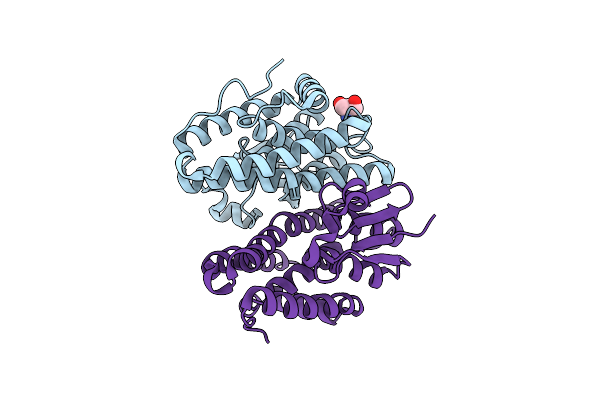 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Apo-Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: TRS |
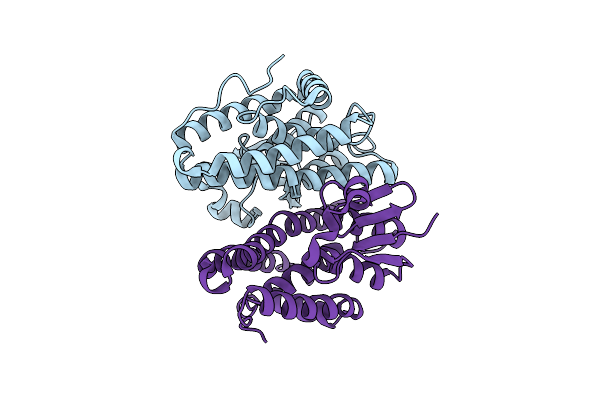 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Apo-Form 2
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-10-02 Classification: TRANSFERASE |
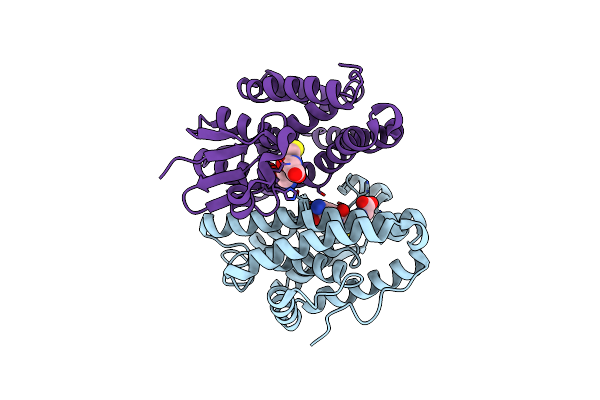 |
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Glutathione-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: GSH |
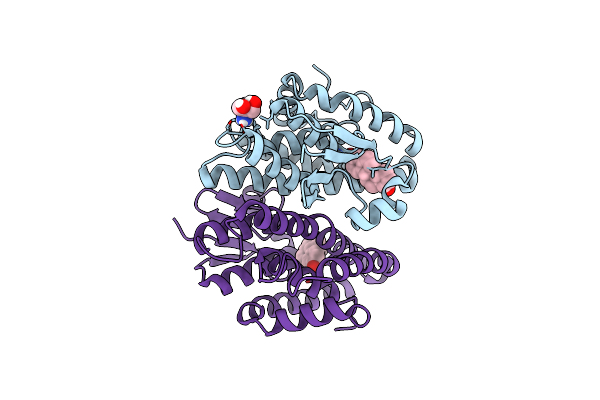 |
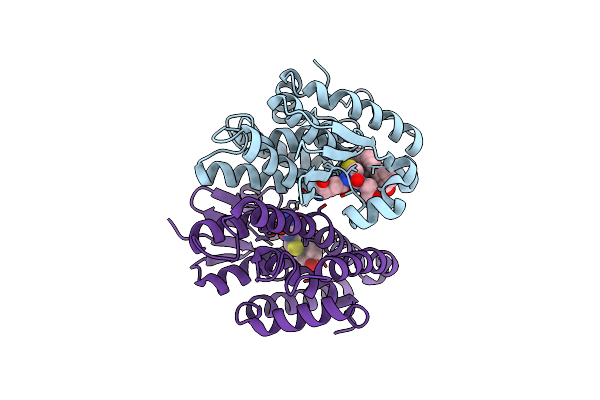
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In 17Beta-Estradiol-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: EST, TRS |
 |
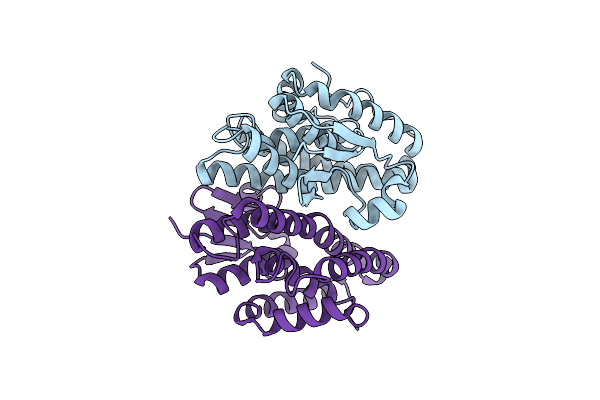
Crystal Structure Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In 17Beta-Estradiol- And Glutathione-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: GSH, EST |
 |
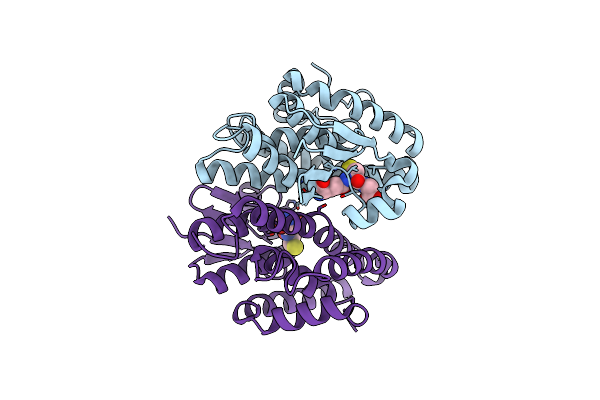
Crystal Structure Of D113A Mutant Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Apo-Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-10-02 Classification: TRANSFERASE |
 |
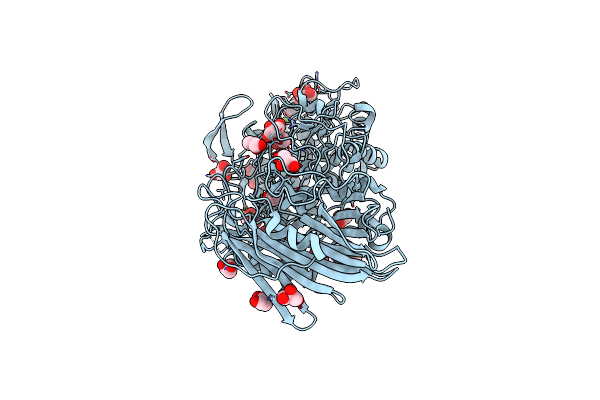
Crystal Structure Of D113A Mutant Of Drosophila Melanogaster Noppera-Bo, Glutathione S-Transferase Epsilon 14 (Dmgste14), In Glutathione-Bound Form
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: GSH |
 |
Crystal Structure Of Thermoanaerobacterium Xylolyticum Gh116 Beta-Glucosidase
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: GOL, CA, EDO |
 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With 2-Deoxy-2-Fluoroglucoside
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: G2F, GOL, EDO, CA |
 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With Deoxynojirimycin
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: NOJ, GOL, CA, EDO |
 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With Glucoimidazole
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: GIM, GOL, CA, EDO |
 |
Crystal Structure Of Thermoanaerobacterium Xylanolyticum Gh116 Beta-Glucosidase With Glucose
Organism: Thermoanaerobacterium xylanolyticum lx-11
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-05-18 Classification: HYDROLASE Ligands: BGC, GOL, EDO, CA |
 |
Bacterial Beta-Glucosidase Reveals The Structural And Functional Basis Of Genetic Defects In Human Glucocerebrosidase 2 (Gba2)
Organism: Thermoanaerobacterium xylanolyticum
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-05-11 Classification: HYDROLASE Ligands: CA |
 |
Rice Os3Bglu6 E178Q With Covalent Glucosyl Moiety From P-Nitrophenyl Glucopyranoside.
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-09-04 Classification: HYDROLASE Ligands: GLC, GOL |
 |
Rice Os3Bglu6 Beta-Glucosidase E178Q Mutant In A Covalent Complex With Glc From Ga4Ge.
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2013-09-04 Classification: HYDROLASE Ligands: GLC, GOL |
 |
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2011-05-18 Classification: HYDROLASE Ligands: TRS, ZN |
 |
The Crystal Structure Of Rice (Oryza Sativa L.) Os4Bglu12 With 2-Fluoroglucopyranoside
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-05-18 Classification: HYDROLASE Ligands: G2F, ZN, GOL |
 |
The Crystal Structure Of Rice (Oryza Sativa L.) Os4Bglu12 With Dinitrophenyl 2-Deoxy-2-Fluoro-Beta-D-Glucopyranoside
Organism: Oryza sativa
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2011-05-18 Classification: HYDROLASE Ligands: GOL, NFG, ZN |

