Search Count: 15
All
Selected
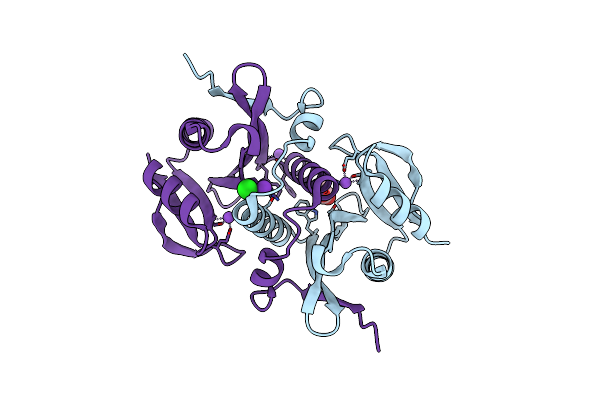 |
Crystal Structure Of Spermin/Spermidine N-Acetyltransferase From Enterococcus Faecalis V583
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-05-28 Classification: TRANSFERASE Ligands: ACT, CL, NA, K |
 |
Poxta-Eq2 Antibiotic Resistance Abcf Bound To E. Faecalis 70S Ribosome, State I
Organism: Enterococcus faecalis v583, Enterococcus faecalis (strain atcc 700802 / v583)
Method: ELECTRON MICROSCOPY Release Date: 2022-03-23 Classification: RIBOSOME Ligands: ATP, MG, ZN, K, PUT, SCM |
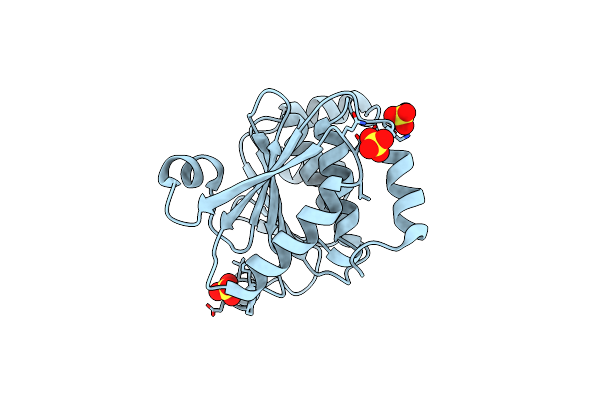 |
Structure Of Enterococcus Faecalis (V583) Alkylhydroperoxide Reductase Subunit F (Ahpf) C503A Mutant
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-05-22 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-09-05 Classification: HYDROLASE Ligands: ZN, PG4, GOL |
 |
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2018-09-05 Classification: HYDROLASE Ligands: CU, DAL, GOL, PG4 |
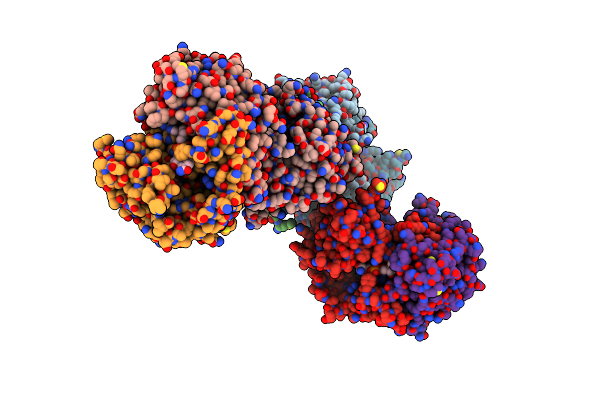 |
Crystal Structure Of Nad Synthetase (Nade) From Enterococcus Faecalis In Complex With Nad+
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2018-02-14 Classification: TRANSFERASE Ligands: NAD |
 |
Mevalonate Diphosphate Mediated Atp Binding Mechanism Of The Mevalonate Diphosphate Decarboxylase From Enterococcus Faecalis
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-10-18 Classification: LYASE Ligands: ATP, PO4 |
 |
Mevalonate Diphosphate Mediated Atp Binding Mechanism Of The Mevalonate Diphosphate Decarboxylase From Enterococcus Faecalis
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-10-18 Classification: LYASE Ligands: SO4 |
 |
Novel Structural Components Contribute To The High Thermal Stability Of Acyl Carrier Protein From Enterococcus Faecalis
Organism: Enterococcus faecalis v583
Method: SOLUTION NMR Release Date: 2015-12-09 Classification: BIOSYNTHETIC PROTEIN |
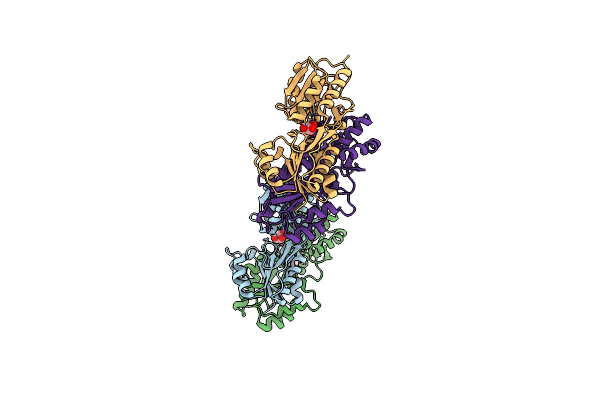 |
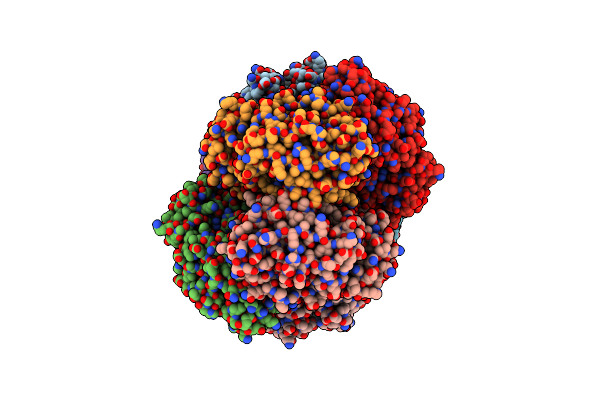
Crystal Structure Of Laci Family Transcriptional Regulator From Enterococcus Faecalis V583, Target Efi-512923, With Bound Ribose
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-10-29 Classification: TRANSCRIPTION REGULATOR Ligands: RIB |
 |
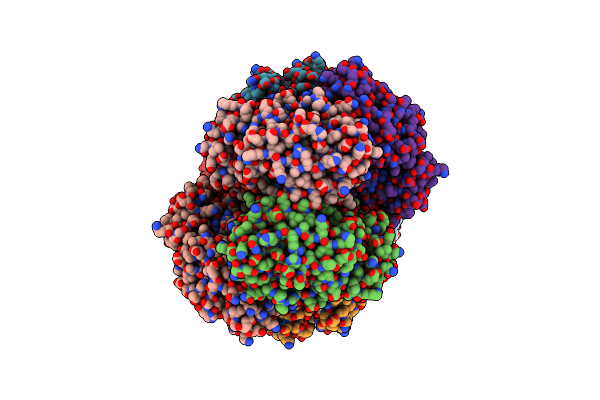
Crystal Structure Of Dipeptide Epimerase From Enterococcus Faecalis V583 Complexed With Mg And Dipeptide L-Leu-L-Tyr
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-08-18 Classification: ISOMERASE Ligands: LEU, TYR, MG |
 |
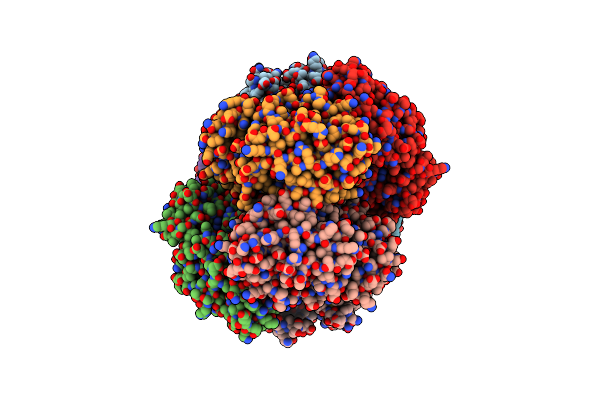
Crystal Structure Of Dipeptide Epimerase From Enterococcus Faecalis V583 Complexed With Mg And Dipeptide L-Ser-L-Tyr
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-08-18 Classification: ISOMERASE Ligands: SER, TYR, GOL, MG |
 |
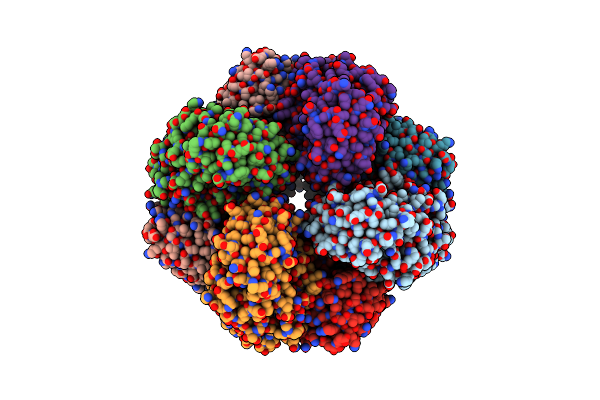
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2010-08-11 Classification: ISOMERASE Ligands: GOL, MG, SO4 |
 |
Crystal Structure Of Dipeptide Epimerase From Enterococcus Faecalis V583 Complexed With Mg And Dipeptide L-Ile-L-Tyr
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-08-11 Classification: ISOMERASE Ligands: ILE, TYR, GOL, MG |
 |
Crystal Structure Of Activating Signal Cointegrator (Np_814290.1) From Enterococcus Faecalis V583 At 1.58 A Resolution
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2009-09-15 Classification: RNA BINDING PROTEIN Ligands: CAC, SO4 |

