Search Count: 1,401
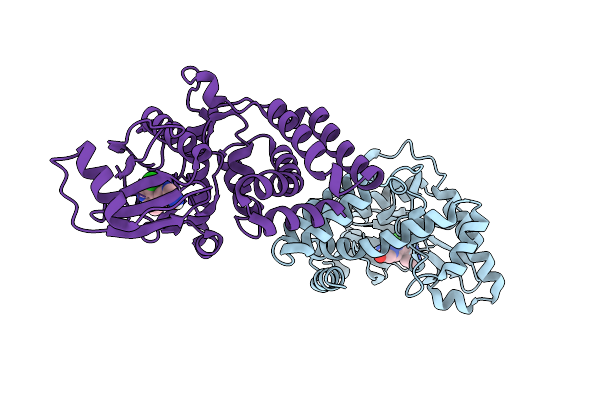 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: ANTIBIOTIC Ligands: DMS, A1I8X |
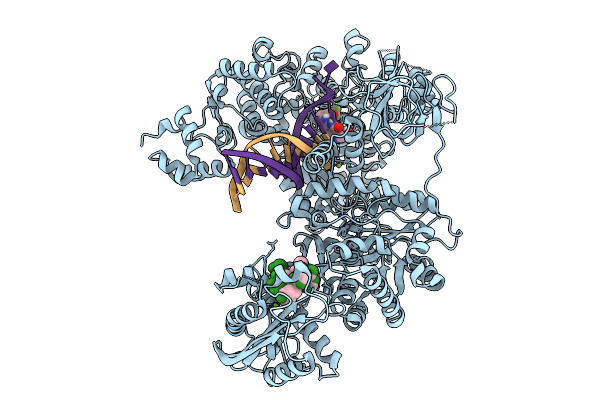 |
Organism: Enterococcus faecium, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DNA BINDING PROTEIN Ligands: A1I88, ZN, MG |
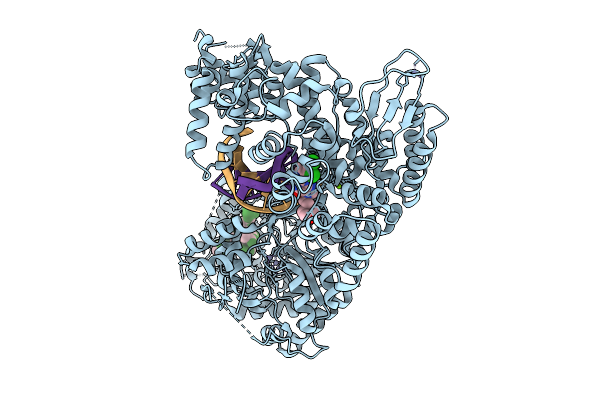 |
Organism: Enterococcus faecium, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DNA BINDING PROTEIN Ligands: ZN, MG, A1I9T |
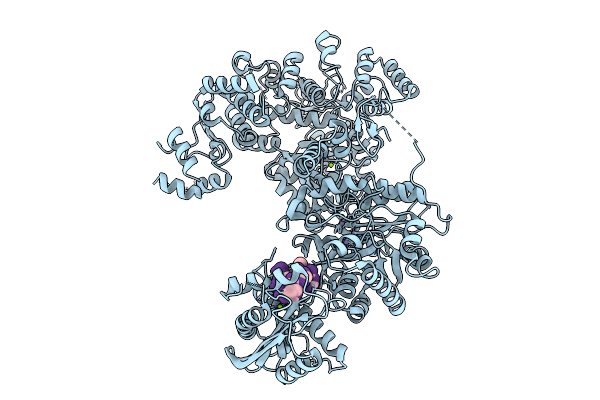 |
Organism: Enterococcus faecium, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DNA BINDING PROTEIN Ligands: ZN, MG |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: METAL BINDING PROTEIN Ligands: MN |
 |
Crystal Structure Of The Peptidyl-Prolyl Isomerase (Ppiase) From E. Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: ISOMERASE Ligands: GOL, CD |
 |
Crystal Structure Of C-Terminal Rigid Fragment Containing Middle And C-Terminal Domains Of The Basal Pilin Ebpb From Enterococcus Faecalis.
Organism: Enterococcus faecalis og1rf
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: CELL ADHESION |
 |
Crystal Structure Of N-Terminal Flexible Domain Of The Shaft Pilin Ebpc From Enterococcus Faecalis.
Organism: Enterococcus faecalis og1rf
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: CELL ADHESION |
 |
Crystal Structure Of N-Terminal Flexible Domain Of The Shaft Pilin Ebpc From Enterococcus Faecalis.
Organism: Enterococcus faecalis og1rf
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: CELL ADHESION |
 |
Organism: Enterococcus faecalis og1rf
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: CELL ADHESION |
 |
Crystal Structure Of C-Terminal Stable Fragment Of The Shaft Pilin Ebpc From Enterococcus Faecalis
Organism: Enterococcus faecalis og1rf
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: CELL ADHESION Ligands: MG, NA |
 |
Crystal Structure Of The Basal Pilin Ebpb From Enterococcus Faecalis With A Partially Disordered N-Terminal Domain.
Organism: Enterococcus faecalis og1rf
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: CELL ADHESION |
 |
Inhibiting The Virulence Of Gut Bacteria Through Blocking The Activation Of A Two-Component Lanthipeptide Toxin
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE |
 |
Organism: Enterococcus faecalis jh1
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: RNA Ligands: K, MG |
 |
Organism: Enterococcus faecalis jh1
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: RNA |
 |
Crystal Structure Of Spermin/Spermidine N-Acetyltransferase From Enterococcus Faecalis V583
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-05-28 Classification: TRANSFERASE Ligands: ACT, CL, NA, K |
 |
Organism: Enterococcus faecalis
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RNA |
 |
Organism: Enterococcus faecalis jh1
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RNA |
 |
Organism: Enterococcus faecalis
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: RNA |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-03-19 Classification: TRANSFERASE Ligands: PLS, HZI |

