Search Count: 31
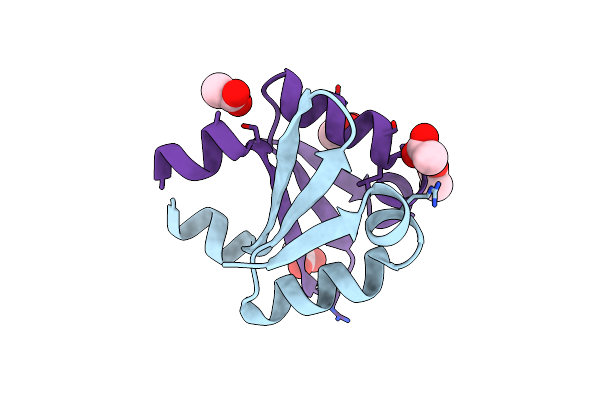 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION Ligands: ACT |
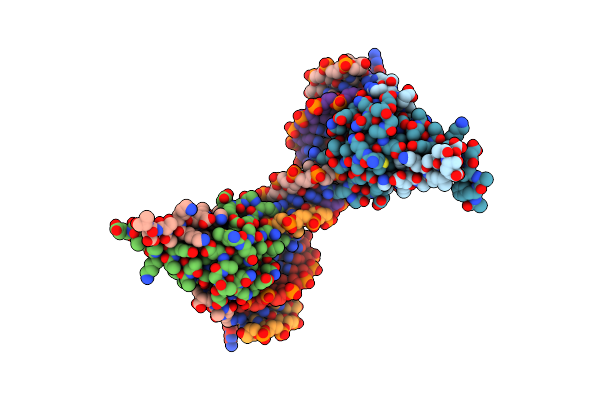 |
Antitoxin Phd From Phage P1 In Complex With Its Operator Dna Inverted Repeat
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.17 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION |
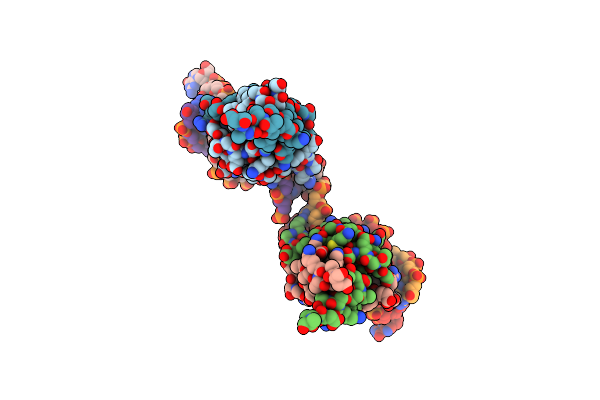 |
Antitoxin Phd From Phage P1 In Complex With Its Operator Dna Inverted Repeat In A Monoclinic Space Group
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.88 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION |
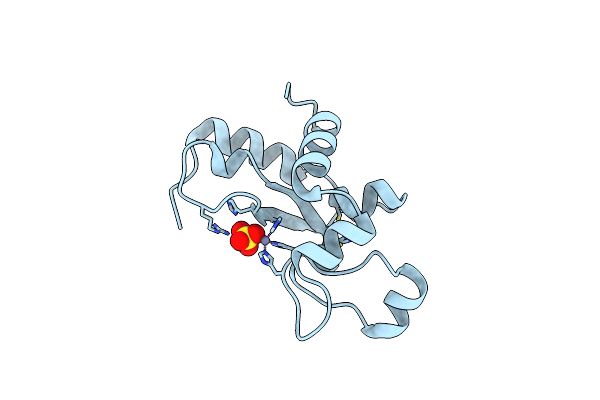 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-12-29 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Organism: Enterobacteria phage p1, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-08-18 Classification: TOXIN/ANTITOXIN Ligands: ACT |
 |
Crystal Structure Of Phd Truncated To Residue 57 In An Orthorhombic Space Group
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-06-23 Classification: ANTITOXIN Ligands: SO4 |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2010-05-26 Classification: ISOMERASE/DNA Ligands: VO4 |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2009-06-09 Classification: RIBOSOME INHIBITOR |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-09-16 Classification: RIBOSOME INHIBITOR Ligands: BR |
 |
Crystal Structure Of The Pre-Cleavage Synaptic Complex In The Cre-Loxp Site-Specific Recombination
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-06-26 Classification: recombination/DNA |
 |
Crystal Structure Of The Tetrameric Pre-Cleavage Synaptic Complex In The Cre-Loxp Site-Specific Recombination
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-06-26 Classification: RECOMBINATION/DNA |
 |
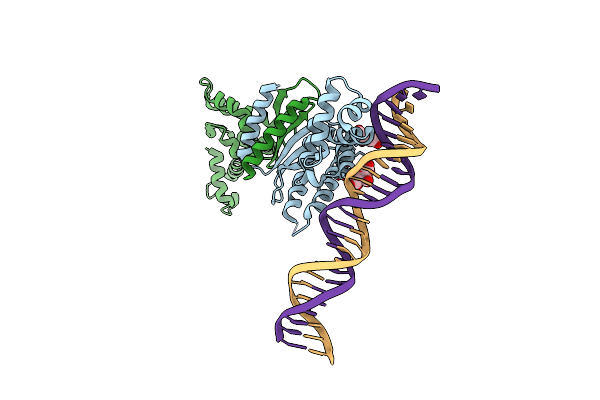
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2007-02-20 Classification: CELL CYCLE/DNA |
 |
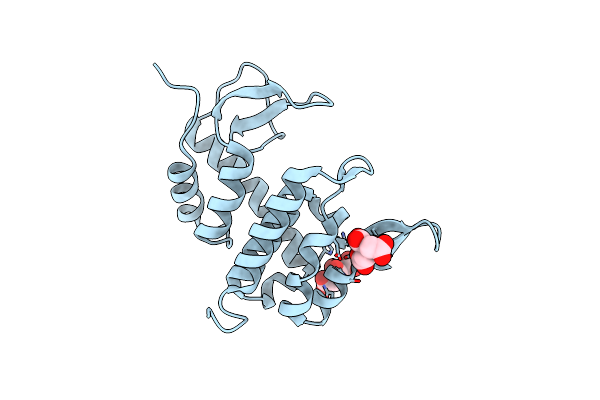
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2005-11-29 Classification: CELL CYCLE Ligands: CIT |
 |
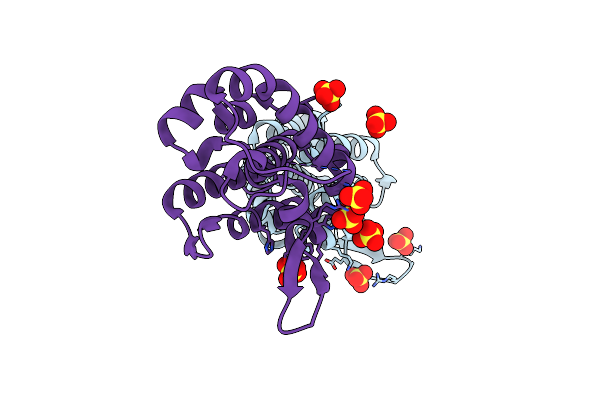
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2005-01-11 Classification: HYDROLASE Ligands: CIT |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2005-01-11 Classification: HYDROLASE Ligands: SO4 |
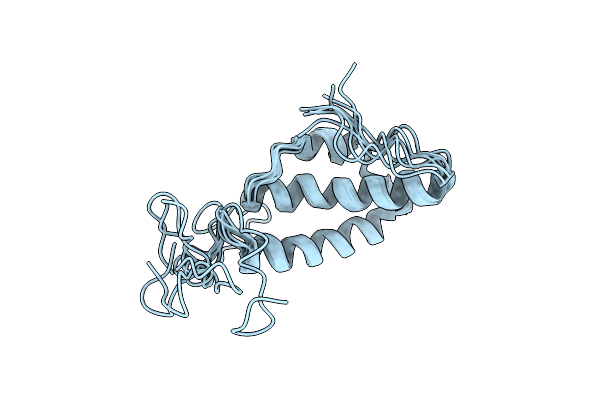 |
Solution Structure Of The E. Coli Bacteriophage P1 Encoded Hot Protein: A Homologue Of The Theta Subunit Of E. Coli Dna Polymerase Iii
Organism: Enterobacteria phage p1
Method: SOLUTION NMR Release Date: 2004-12-14 Classification: TRANSFERASE |
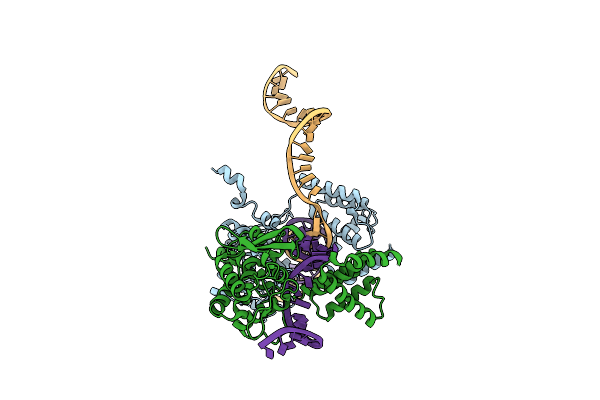 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-12-14 Classification: Hydrolase, Ligase/DNA |
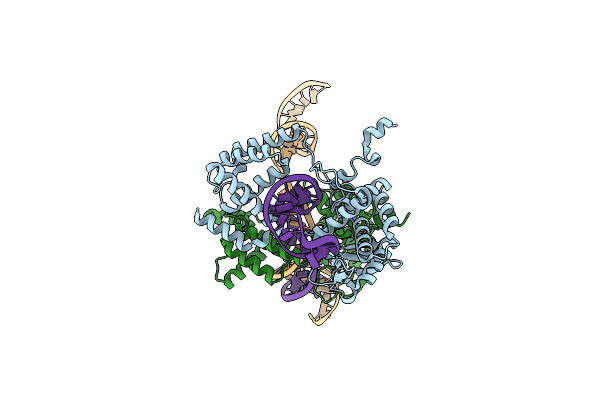 |
High Resolution Structure Of The Holliday Junction Intermediate In Cre-Loxp Site-Specific Recombination
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-12-14 Classification: HYDROLASE, LIGASE/DNA |
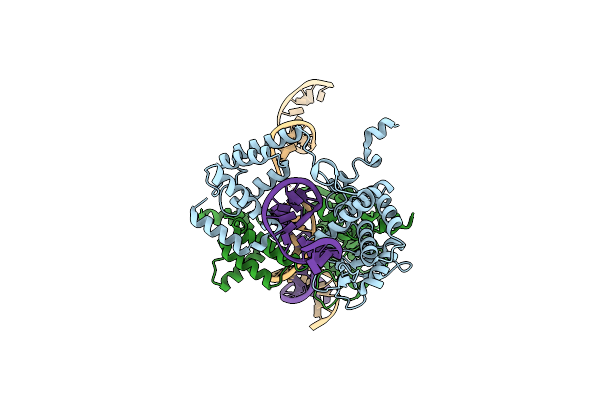 |
Basis For A Switch In Substrate Specificity: Crystal Structure Of Selected Variant Of Cre Site-Specific Recombinase, Lnsgg Bound To The Loxp (Wildtype) Recognition Site
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2004-02-17 Classification: RECOMBINATION/DNA |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2003-09-16 Classification: REPLICATION/DNA Ligands: MG, IOD |

