Search Count: 29
 |
Cryo- Em Structure Of Large Subunit (Lsu) Of 75S Ribosome With P- Trna From Entamoeba Histolytica
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: RIBOSOME |
 |
Cryo- Em Structure Of 75S Ribosome With A/P- & P/E- Trnas From Entamoeba Histolytica
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: RIBOSOME |
 |
Cryo- Em Structure Of Large Subunit (Lsu) Of 75S Ribosome With A/P- & P/E- Trnas From Entamoeba Histolytica
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: RIBOSOME |
 |
Cryo- Em Structure Of Ribosomal Large Subunit (Lsu) From Entamoeba Histolytica At 2.8 Angstrom Resolution
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: RIBOSOME |
 |
Cryo- Em Structure Of Small Subunit (Body) Of 75S Ribosome With P- Trna From Entamoeba Histolytica
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: RIBOSOME |
 |
Cryo- Em Structure Of Small Subunit (Head) Of 75S Ribosome With P- Trna From Entamoeba Histolytica
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: RIBOSOME |
 |
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: RIBOSOME |
 |
Cryo- Em Structure Of 75S Ribosome With A/P- & P/E- Trnas From Entamoeba Histolytica Bound To Antibiotic Paromomycin
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: RIBOSOME Ligands: PAR |
 |
Cryo- Em Structure Of Small Subunit (Body) Of 75S Ribosome With A/P- & P/E- Trnas From Entamoeba Histolytica
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: RIBOSOME |
 |
Cryo- Em Structure Of Small Subunit (Head) Of 75S Ribosome With A/P- & P/E- Trnas From Entamoeba Histolytica
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: RIBOSOME |
 |
Cryo- Em Structure Of Small Subunit (Body) Of 75S Ribosome With A/P- & P/E- Trnas From Entamoeba Histolytica Bound To Antibiotic Paromomycin
Organism: Entamoeba histolytica hm-1:imss
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: RIBOSOME Ligands: PAR |
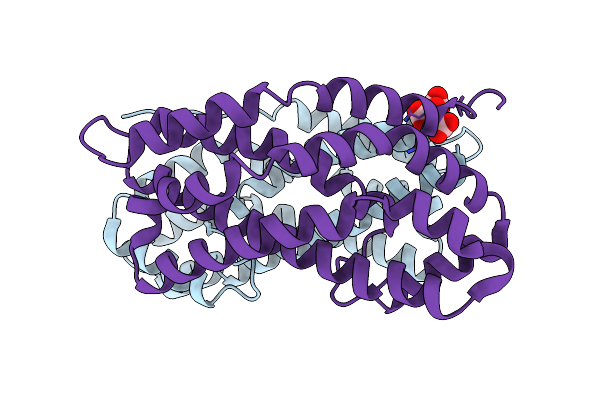 |
Crystal Structure Of Dh Domain Of Fyve Domain Containing Protein(Fp10) From Entamoeba Histolytica
Organism: Entamoeba histolytica hm-1:imss-a
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2025-05-21 Classification: SIGNALING PROTEIN Ligands: CIT |
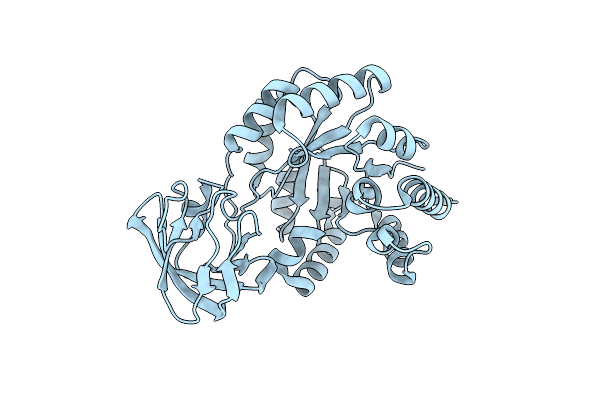 |
Organism: Entamoeba histolytica hm-1:imss
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-04-10 Classification: CYTOSOLIC PROTEIN |
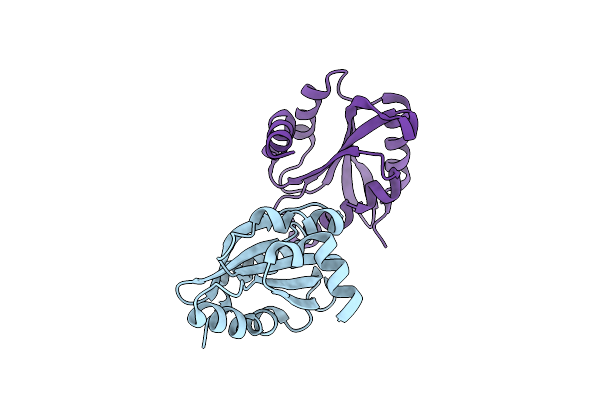 |
Organism: Entamoeba histolytica hm-1:imss
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-06-03 Classification: STRUCTURAL PROTEIN |
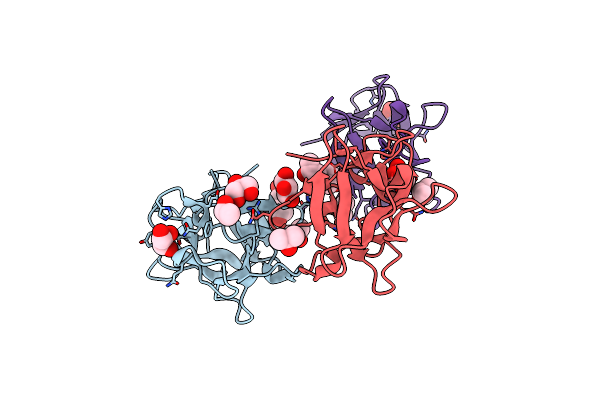 |
Organism: Entamoeba histolytica hm-1:imss
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-09-25 Classification: SUGAR BINDING PROTEIN Ligands: GOL, IPA |
 |
Organism: Entamoeba histolytica hm-1:imss
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2019-09-25 Classification: SUGAR BINDING PROTEIN Ligands: RM4, GOL, IPA |
 |
Crystal Structure Of Phosphoserine Phosphatase Mutant (H9A) From Entamoeba Histolytica In Complex With Phosphoserine
Organism: Entamoeba histolytica hm-1:imss-a
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2019-04-24 Classification: HYDROLASE Ligands: SEP, SO4, PG4, PEG |
 |
Organism: Entamoeba histolytica hm-1:imss-a
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-04-03 Classification: HYDROLASE Ligands: BME, PO4, PEG, 144, GOL, PG4 |
 |
Organism: Entamoeba histolytica hm-1:imss-a
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-02-20 Classification: UNKNOWN FUNCTION |
 |
The Crystal Structure Of The Two Spectrin Repeat Domains From Entamoeba Histolytica
Organism: Entamoeba histolytica hm-1:imss
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2018-05-16 Classification: STRUCTURAL PROTEIN Ligands: BME |

