Search Count: 28
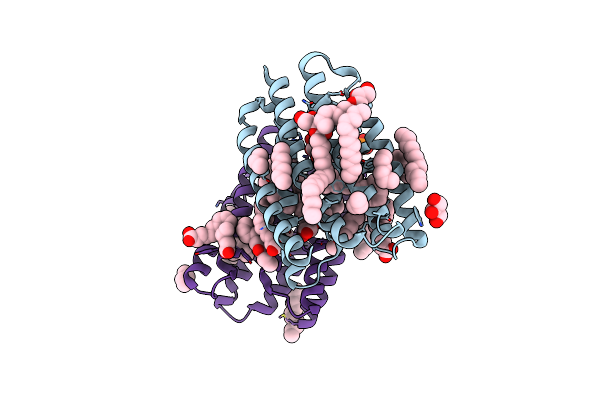 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
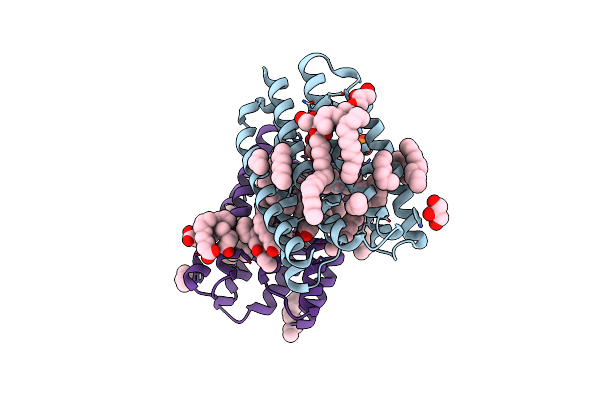 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
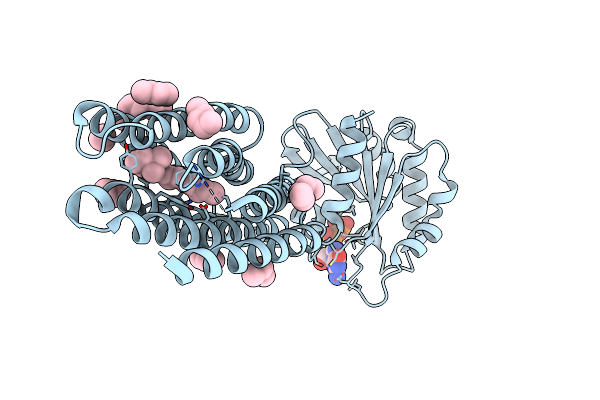 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
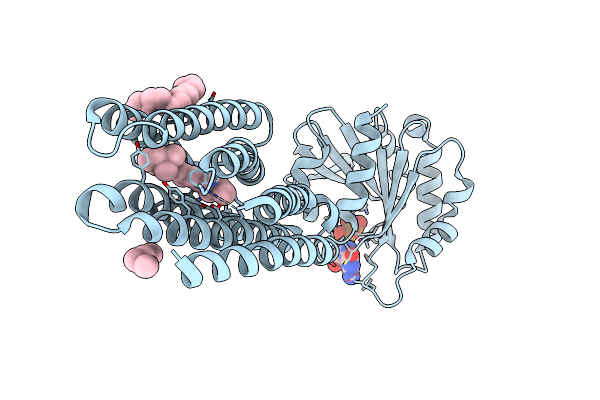 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State Obtained By Cryotrapping
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Synechocystis Halorhodopsin (Syhr), Cl-Pumping Mode, Ground State
Organism: Synechocystis sp. pcc 7509
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2022-11-09 Classification: MEMBRANE PROTEIN Ligands: CL, LFA, OLC, OLA |
 |
Crystal Structure Of Synechocystis Halorhodopsin (Syhr), Cl-Pumping Mode, K State
Organism: Synechocystis sp. pcc 7509
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-11-09 Classification: MEMBRANE PROTEIN Ligands: CL, LFA, OLC, OLA |
 |
Crystal Structure Of Synechocystis Halorhodopsin (Syhr), Cl-Pumping Mode, O State
Organism: Synechocystis sp. pcc 7509
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-09 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA, OLC |
 |
Crystal Structure Of Synechocystis Halorhodopsin (Syhr), So4-Bound Form, Ground State
Organism: Synechocystis sp. pcc 7509
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-11-09 Classification: MEMBRANE PROTEIN Ligands: CL, SO4, LFA, OLA, GOL |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The M-Like State
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-06-01 Classification: MEMBRANE PROTEIN Ligands: LFA, OLB, OLC, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2022-06-01 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Crystal Structure Of The K State Of Bacteriorhodopsin At 1.53 Angstrom Resolution
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-05-18 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC, OLA |
 |
Crystal Structure Of The M State Of Bacteriorhodopsin At 1.22 Angstrom Resolution
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2022-05-18 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, L2P, SQL |
 |
Crystal Structure Of The Ground State Of Bacteriorhodopsin At 1.05 Angstrom Resolution
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2022-05-04 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC, OLA |
 |
Crystal Structure Of The Ground State Of Bacteriorhodopsin At 1.22 Angstrom Resolution
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2022-05-04 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC, OLA |
 |
Crystal Structure Of The L State Of Bacteriorhodopsin At 1.20 Angstrom Resolution
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2022-05-04 Classification: MEMBRANE PROTEIN Ligands: LFA, OLC, OLA |
 |
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-02-15 Classification: SIGNALING PROTEIN Ligands: RET, BOG, LFA |
 |
Structure Of The Srii/Htrii Complex In I212121 Space Group ("U" Shape) - M State
Organism: Natronomonas pharaonis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-02-15 Classification: SIGNALING PROTEIN Ligands: RET, BOG, LFA |

