Search Count: 19
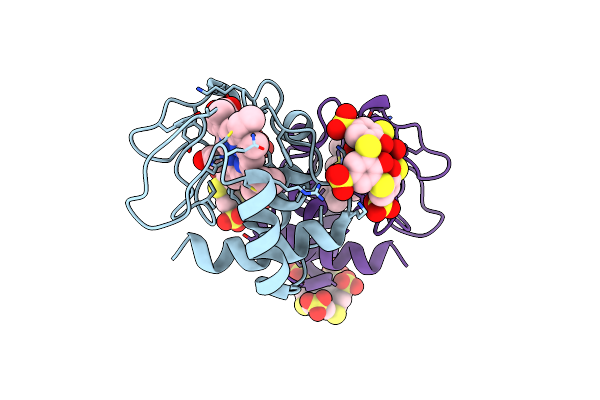 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-03-02 Classification: ELECTRON TRANSPORT Ligands: HEC, 80M |
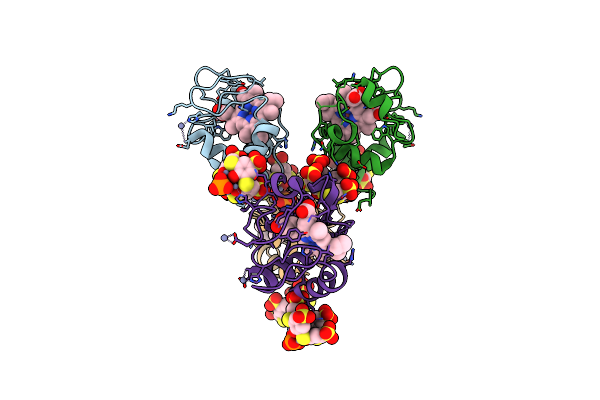 |
Cocrystal Form I Of A Cytochrome C, Sulfonato-Thiacalix[4]Arene - Zinc Cluster
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Release Date: 2022-03-02 Classification: ELECTRON TRANSPORT Ligands: HEC, ZN, 80M, PO4 |
 |
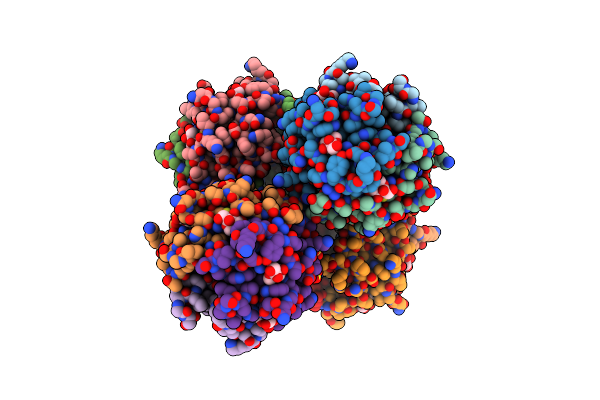
Cocrystal Form Ii Of A Cytochrome C, Sulfonato-Thiacalix[4]Arene - Zinc Cluster
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2022-03-02 Classification: ELECTRON TRANSPORT Ligands: HEC, 80M, ZN, PO4 |
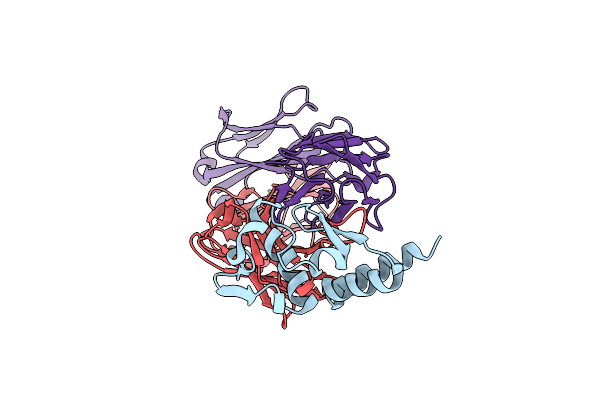
 |
Organism: Ralstonia solanacearum
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-03-02 Classification: SUGAR BINDING PROTEIN Ligands: BDF, ZN, GOL, 80M |
 |
Crystal Structure Of C-Terminal Domain Of Ebola (Reston) Nucleoprotein In Complex With Fab Fragment
Organism: Homo sapiens, Reston ebolavirus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2018-06-13 Classification: viral protein/immune system |
 |
Crystal Structure Of C-Terminal Domain Of Ebola (Bundibugyo) Nucleoprotein In Complex With Fab Fragment
Organism: Bundibugyo ebolavirus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-04-25 Classification: IMMUNE SYSTEM |
 |
Organism: Zika virus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2017-09-06 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
Organism: Zika virus (strain mr 766)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-09-06 Classification: VIRAL PROTEIN Ligands: SAM, SO4 |
 |
The Crystal Structure Of The C-Terminal Domain Of Ebola (Tai Forest) Nucleoprotein
Organism: Tai forest ebolavirus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-10-21 Classification: VIRAL PROTEIN Ligands: 2PE |
 |
The Crystal Structure Of The C-Terminal Domain Of Ebola (Bundibugyo) Nucleoprotein
Organism: Bundibugyo virus
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2015-09-30 Classification: VIRAL PROTEIN Ligands: GOL, CL |
 |
The Crystal Structure Of The C-Terminal Domain Of Ebola (Zaire) Nucleoprotein
Organism: Ebola virus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2014-09-10 Classification: VIRAL PROTEIN |
 |
The Crystal Structure Of The C-Terminal Domain Of Ebola (Zaire) Nucleoprotein
Organism: Ebola virus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-09-10 Classification: VIRAL PROTEIN |
 |
Method: X-RAY DIFFRACTION
Resolution:2.70 Å Release Date: 2005-03-22 Classification: DE NOVO PROTEIN Ligands: CD |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2005-03-01 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2005-03-01 Classification: DE NOVO PROTEIN Ligands: ZN |
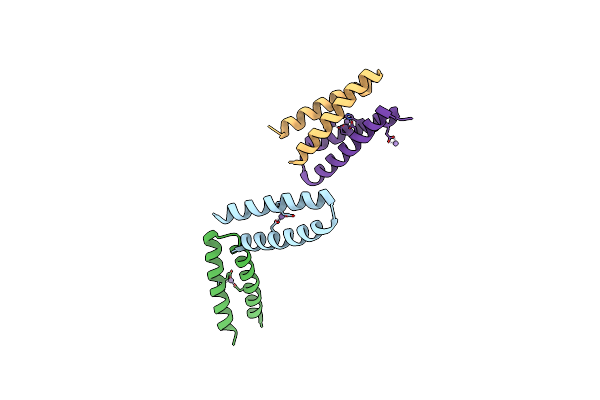 |
Method: X-RAY DIFFRACTION
Resolution:2.99 Å Release Date: 2004-05-18 Classification: DE NOVO PROTEIN Ligands: MN |
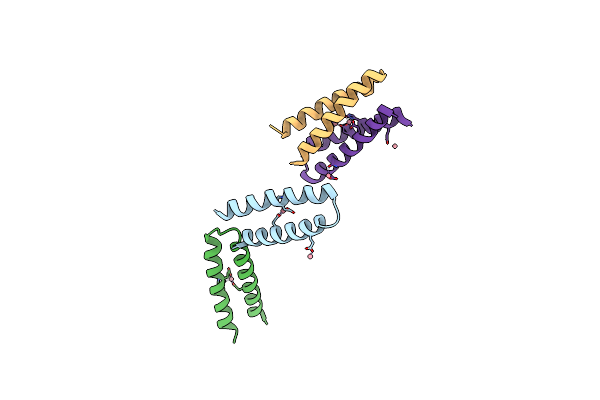 |
Method: X-RAY DIFFRACTION
Resolution:3.10 Å Release Date: 2004-04-06 Classification: DE NOVO PROTEIN Ligands: CO |
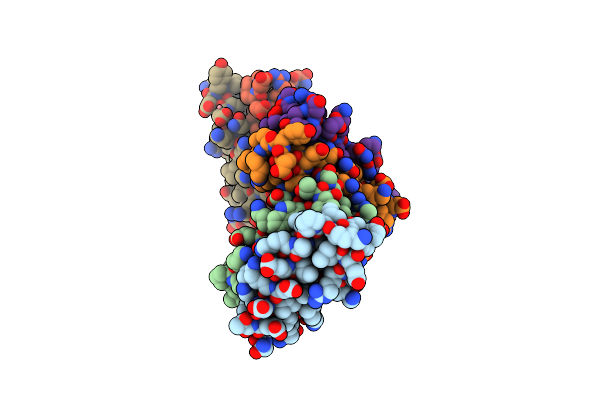 |
Method: X-RAY DIFFRACTION
Resolution:2.90 Å Release Date: 2004-04-06 Classification: DE NOVO PROTEIN Ligands: CO |
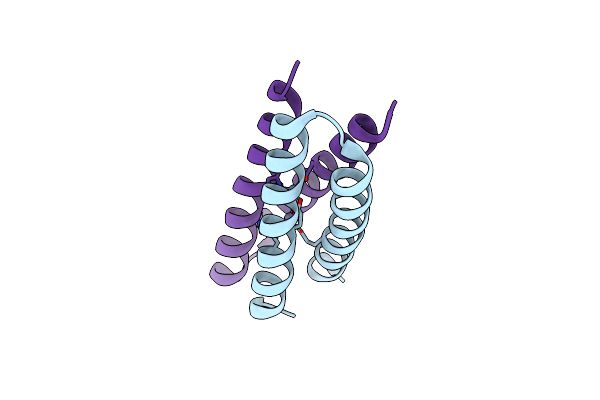 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2004-01-20 Classification: DE NOVO PROTEIN Ligands: ZN |

