Search Count: 46
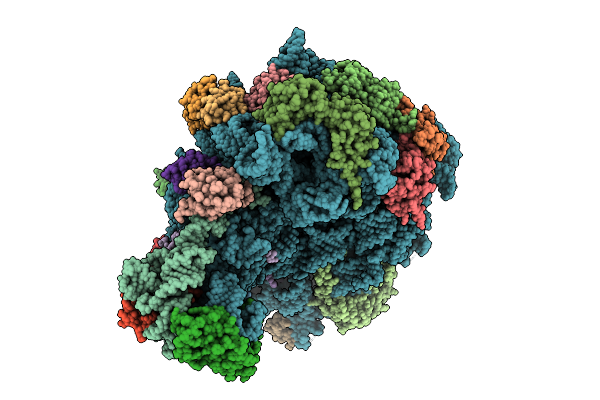 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
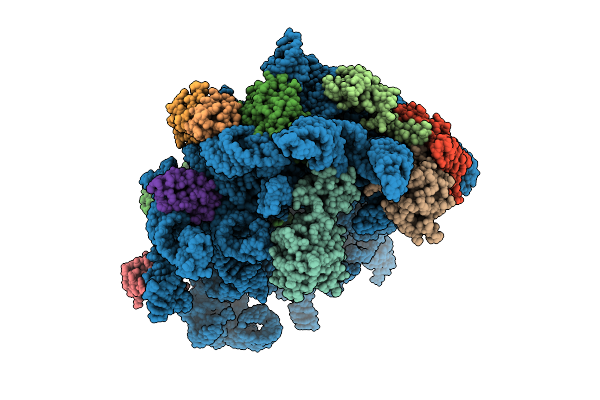 |
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME |
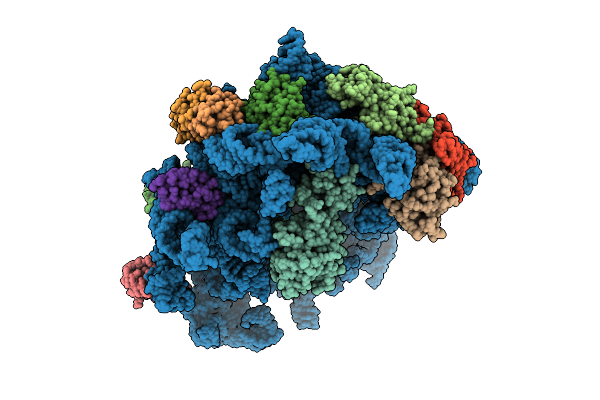 |
Organism: Escherichia coli k-12, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: AN6 |
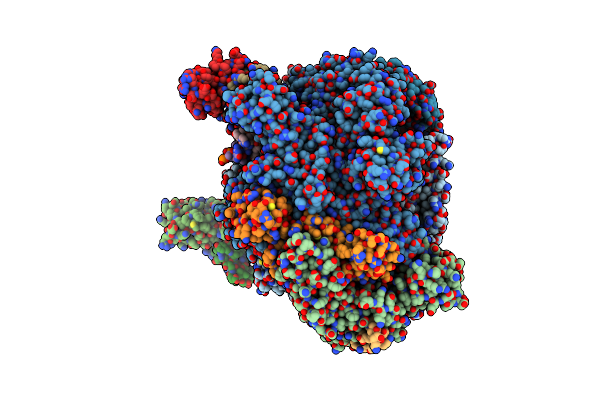 |
Cryo-Em Structure Of Pyrococcus Furiosus Transcription Elongation Complex Bound To Spt4/5
Organism: Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Cryo-Em Structure Of Pyrococcus Furiosus Apo Form Rna Polymerase Open Clamp Conformation
Organism: Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo-Em Structure Of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation With Spt4/5
Organism: Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Cryo-Em Structure Of Pyrococcus Furiosus Apo Form Rna Polymerase Contracted Clamp Conformation
Organism: Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Organism: Pyrococcus furiosus dsm 3638
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: TRANSCRIPTION |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: IMMUNE SYSTEM Ligands: ADP, 8GI |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-05-26 Classification: OXIDOREDUCTASE Ligands: N55, PGE, EDO, HEM |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-05-26 Classification: OXIDOREDUCTASE Ligands: EDO, N5Z, PGE, HEM |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-05-26 Classification: OXIDOREDUCTASE Ligands: N5W, EDO, HEM |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843)
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Schizosaccharomyces pombe (strain 972 / atcc 24843), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-02-24 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2020-12-30 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-12-30 Classification: TRANSCRIPTION Ligands: ACT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-12-23 Classification: TRANSCRIPTION |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-03-18 Classification: TRANSCRIPTION Ligands: ZN, MG |

