Search Count: 357
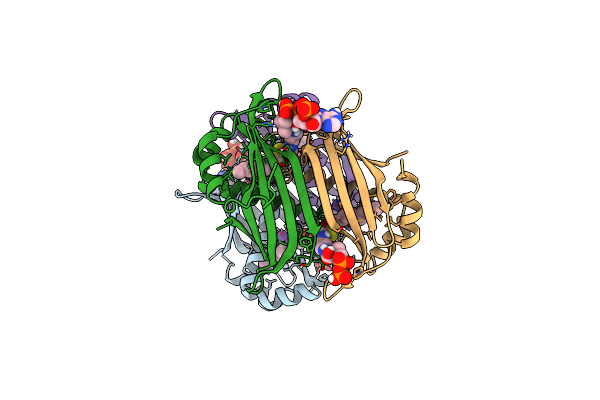 |
Crystal Structure Of The Espe7 Thioesterase Mutant R35A From The Esperamicin Biosynthetic Pathway At 1.6 A
Organism: Actinomadura verrucosospora
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE Ligands: K, DCC |
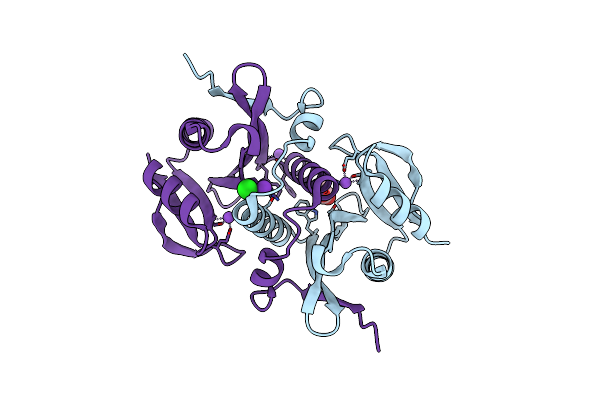 |
Crystal Structure Of Spermin/Spermidine N-Acetyltransferase From Enterococcus Faecalis V583
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-05-28 Classification: TRANSFERASE Ligands: ACT, CL, NA, K |
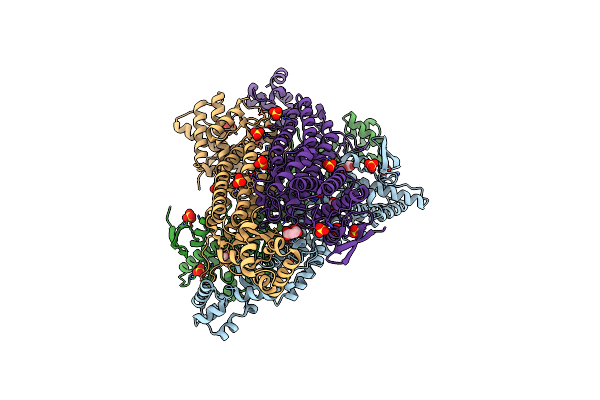 |
Organism: Staphylococcus aureus subsp. aureus usa300_fpr3757
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-14 Classification: HYDROLASE Ligands: FUM, FMT, SO4, CL |
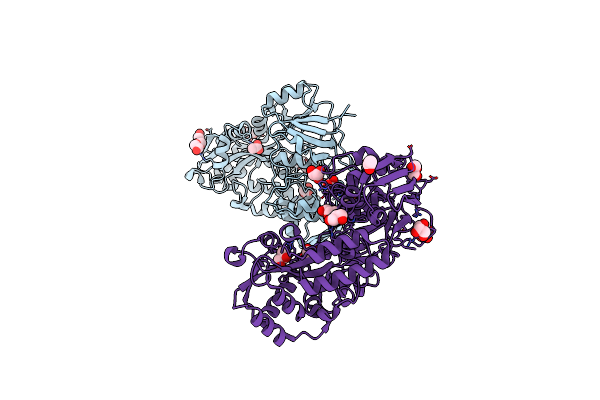 |
Crystal Structure Of Bacterial Extracellular Solute-Binding Protein From Bordetella Bronchiseptica Rb50
Organism: Bordetella bronchiseptica
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-12-18 Classification: STRUCTURAL GENOMICS Ligands: GOL, ACT, PGE, PG4 |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2024-10-09 Classification: HYDROLASE Ligands: EDO, SO4 |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, L-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-10-02 Classification: LYASE Ligands: 8FL, UQ3, ADP, ACE, MG, GOL |
 |
Crystal Structure Of 2-Hydroxacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, D-Lactyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-10-02 Classification: LYASE Ligands: MG, 8FL, UQ3, ADP, EDO, ACY, PO4, CL, ACE, PEG |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthse Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, Formyl-Coa, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-10-02 Classification: LYASE Ligands: A1AEK, FYN, MG, ADP, EDO, PO4, PEG |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Tbhacs From Thermoflexaceae Bacterium In The Complex With Thdp, Formyl-Coa And Adp
Organism: Thermoflexaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-10-02 Classification: LYASE Ligands: TPP, FYN, CA, ADP, EDO, PEG |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Tbhacs From Thermoflexaceae Bacterium In The Complex With Thdp, 2-Hydroxyisobutyryl-Coa And Adp
Organism: Thermoflexaceae bacterium
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-10-02 Classification: LYASE Ligands: A1AEK, COA, ADP, MG, EDO |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Apbhacs From Alphaproteobacteria Bacterium In The Complex With Thdp, Coenzyme A, And Adp
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2024-10-02 Classification: LYASE Ligands: FYN, TPP, MG, ADP, EDO, SO4, CL, GOL |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Tbhacs From Thermoflexaceae Bacterium In The Complex With Thdp And Adp
Organism: Thermoflexaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2024-10-02 Classification: LYASE Ligands: ADP, TPP, EDO, GOL, PO4, MG, CL |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Cchacs From Conidiobolus Coronatus In The Complex With Thdp And Adp
Organism: Conidiobolus coronatus nrrl 28638
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-10-02 Classification: LYASE Ligands: TPP, ADP, EDO, ALA, ACT, MG, CL, GLY, FMT |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Dhchacs From Dehalococcoidia Bacterium In The Complex With Thdp And Adp
Organism: Dehalococcoidia bacterium
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-10-02 Classification: LYASE Ligands: TPP, ADP, GOL, MG, EDO |
 |
Crystal Structure Of 2-Hydroxyacyl-Coa Lyase/Synthase Cfhhacs From Chloroflexi Bacterium In The Complex With Thdp And Adp
Organism: Chloroflexi bacterium hgw-chloroflexi-9
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-10-02 Classification: LYASE Ligands: TPP, ADP, MG, EDO, PEG |
 |
The Crystal Structure Of Geranyltranstransferase From Streptococcus Pneumoniae Tigr4
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2024-09-18 Classification: TRANSFERASE Ligands: GOL |
 |
Crystal Structure Of Lpxtg-Motif Cell Wall Anchor Domain Protein Mscramm_Sdrd From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus jh1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-08-07 Classification: CELL ADHESION Ligands: GOL, EDO, CA |
 |
Crystal Structure Of The 3-Ketoacyl-(Acyl-Carrier-Protein) Reductase, Cylg, From Streptococcus Agalactiae 2603V/R
Organism: Streptococcus agalactiae 2603v/r
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-07-24 Classification: OXIDOREDUCTASE Ligands: NDP, IMD |
 |
Crystal Structure Of Metallo-Beta-Lactamase Superfamily Protein Ccra-Like_Mbl-B1 From Dyadobacter Fermentans
Organism: Dyadobacter fermentans dsm 18053
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-07-17 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of Zn-Dependent Hydrolase From Salmonella Typhimurium Lt2
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-07-10 Classification: HYDROLASE Ligands: EDO, SO4 |

