Search Count: 44
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 8.5 - Apo Form
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: CL, MG |
 |
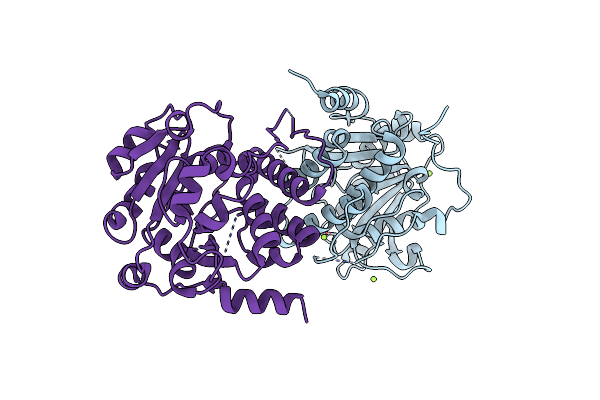
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 In Complex With Udp-Glucose
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-02-22 Classification: TRANSFERASE Ligands: UPG, MG, CL, BCT |
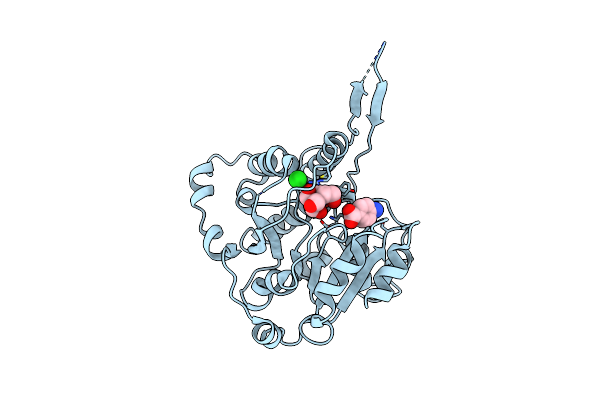 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 In Complex With 4-Aminobenzoic Acid
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-02-15 Classification: TRANSFERASE Ligands: PAB, BTB, CL |
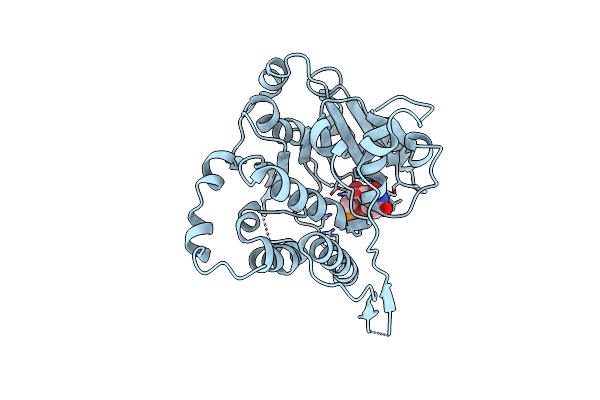 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 In Complex With Udp
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-02-15 Classification: TRANSFERASE Ligands: CL, UDP |
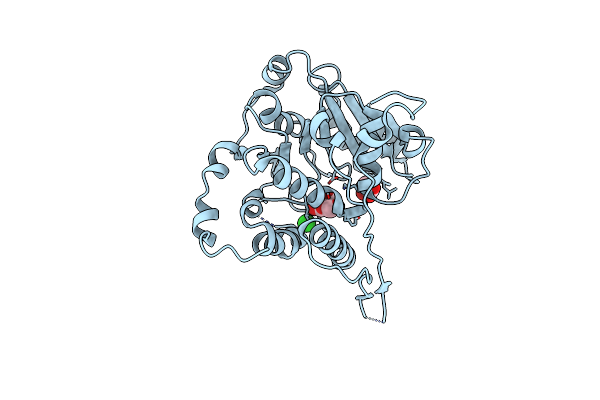 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 - Apo Form
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: CL, BGC, MLI |
 |
Organism: Mycolicibacterium hassiacum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-01-25 Classification: TRANSFERASE |
 |
Organism: Mycolicibacterium hassiacum
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-01-25 Classification: HYDROLASE Ligands: MG, NI, GOL |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.1 - Apo Form
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: LMR, MLT |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 8.5 In Complex With Ump And Magnesium
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: MG, U5P, CL |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.2 - Apo Form
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2022-12-14 Classification: TRANSFERASE Ligands: TLA |
 |
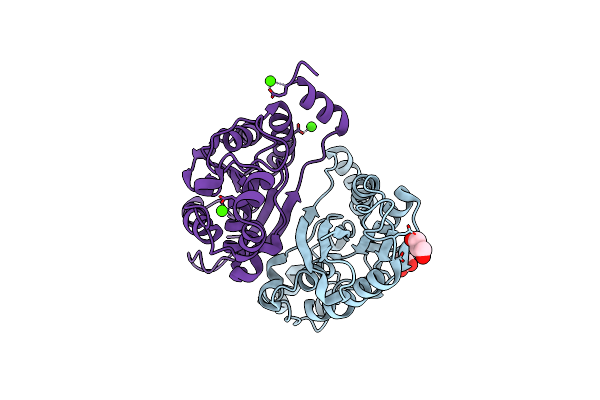
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-07-22 Classification: HYDROLASE Ligands: IOD |
 |
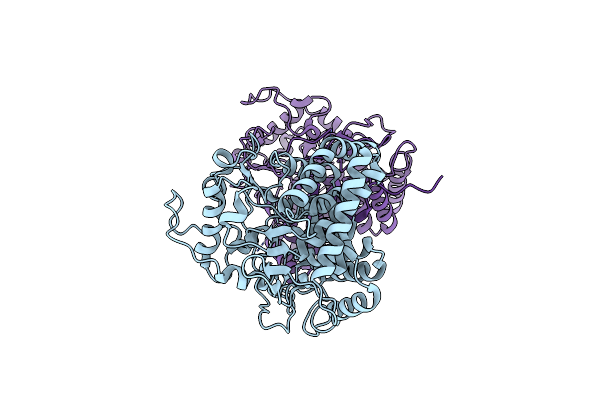
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: GOL, CA |
 |
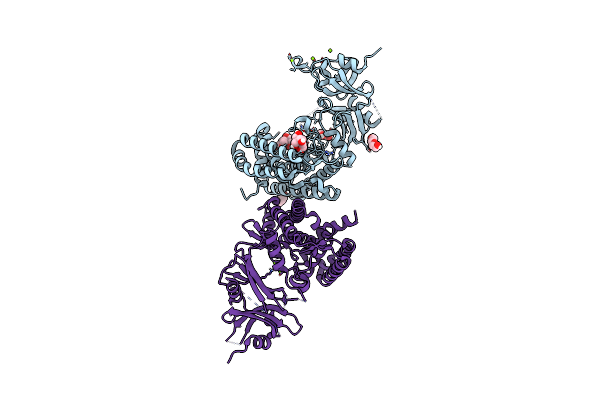
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) - Apo Form
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2019-05-15 Classification: HYDROLASE |
 |
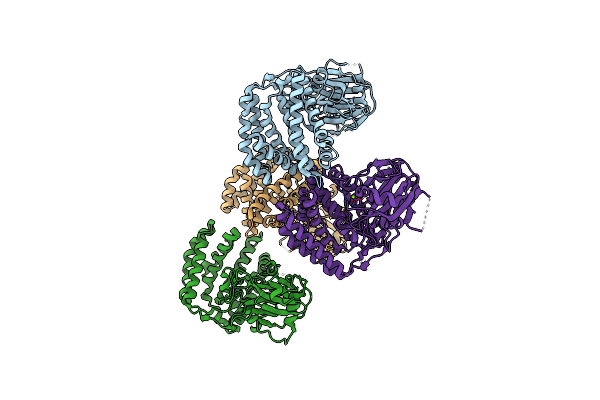
Organism: Streptacidiphilus jiangxiensis
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2019-05-01 Classification: TRANSFERASE Ligands: MG, BTB, PEG, CL |
 |
Organism: Streptacidiphilus jiangxiensis
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2019-05-01 Classification: TRANSFERASE Ligands: CL, MG |
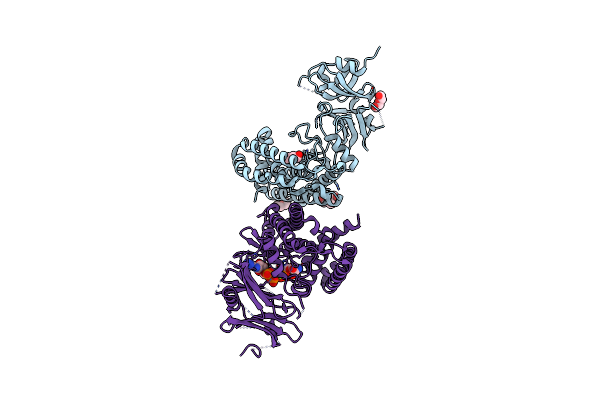 |
Glucosamine Kinase In Complex With Glucosamine, Adp And Inorganic Phosphate
Organism: Streptacidiphilus jiangxiensis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-05-01 Classification: TRANSFERASE Ligands: CL, BTB, PEG, ADP, MG, PO4, GCS |
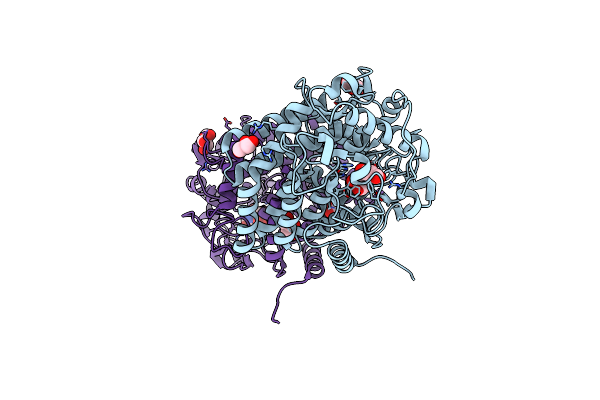 |
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: GOL, SER |
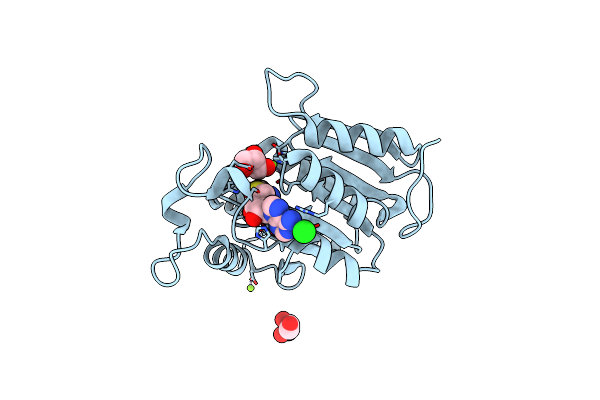 |
Structure Of Met1 From Mycobacterium Hassiacum In Complex With Sam And Glycerol.
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: SAH, GOL, MG, CL |
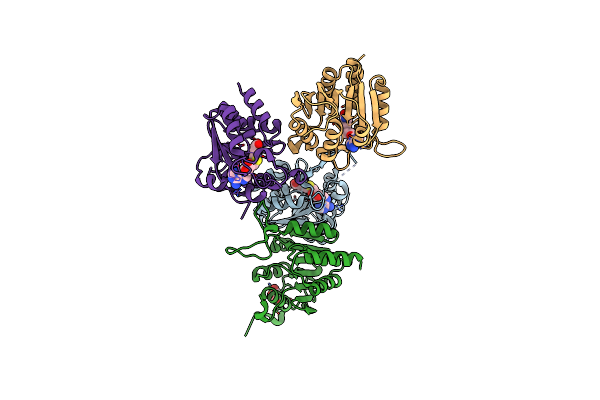 |
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: SAH, GOL |
 |
High Resolution Structure Of Met1 From Mycobacterium Hassiacum In Complex With 3-Methoxy-1,2-Propanediol.
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: GOL, FW5, SAH, MG, CL |

