Search Count: 16
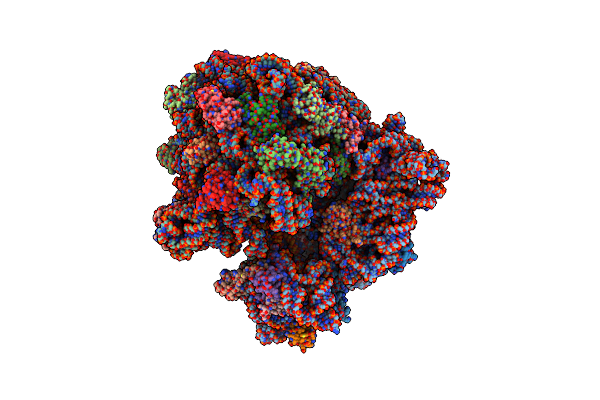 |
Cryo-Em Structure Of The E.Coli 70S Ribosome In Complex With The Antibiotic Myxovalargin B.
Organism: Myxococcus fulvus, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-01-25 Classification: RIBOSOME Ligands: MG, ZN, FME, SPD |
 |
Cryo-Em Structure Of The E.Coli 50S Ribosomal Subunit In Complex With The Antibiotic Myxovalargin A.
Organism: Myxococcus fulvus, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Acinetobacter baumannii (strain atcc 17978 / cip 53.77 / lmg 1025 / ncdc kc755 / 5377)
Method: ELECTRON MICROSCOPY Release Date: 2022-02-02 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP, PO4 |
 |
Organism: Acinetobacter baumannii atcc 17978
Method: ELECTRON MICROSCOPY Release Date: 2022-02-02 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Acinetobacter baumannii (strain atcc 17978 / cip 53.77 / lmg 1025 / ncdc kc755 / 5377)
Method: ELECTRON MICROSCOPY Release Date: 2022-02-02 Classification: MEMBRANE PROTEIN Ligands: ATP, MG, ADP |
 |
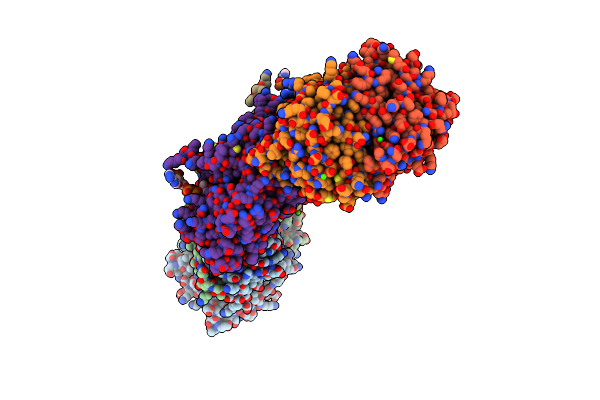
Molecular Basis Of The Flavin-Based Electron-Bifurcating Caffeyl-Coa Reductase Reaction
Organism: Acetobacterium woodii (strain atcc 29683 / dsm 1030 / jcm 2381 / kctc 1655 / wb1)
Method: X-RAY DIFFRACTION Release Date: 2018-01-24 Classification: FLAVOPROTEIN Ligands: FAD, SF4, SO4 |
 |
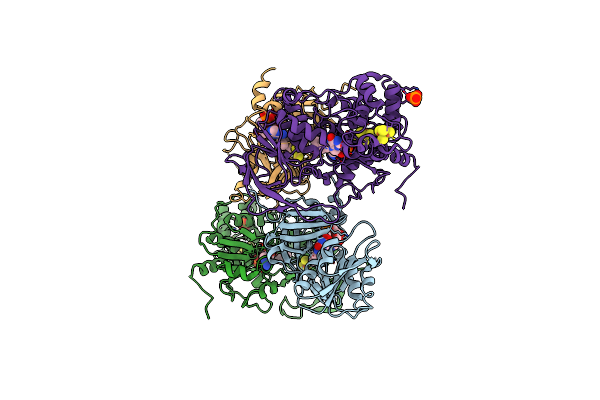
The Electron Transferring Flavoprotein/Butyryl-Coa Dehydrogenase Complex From Clostridium Difficile
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Release Date: 2017-11-29 Classification: FLAVOPROTEIN Ligands: FAD, CA, COS |
 |
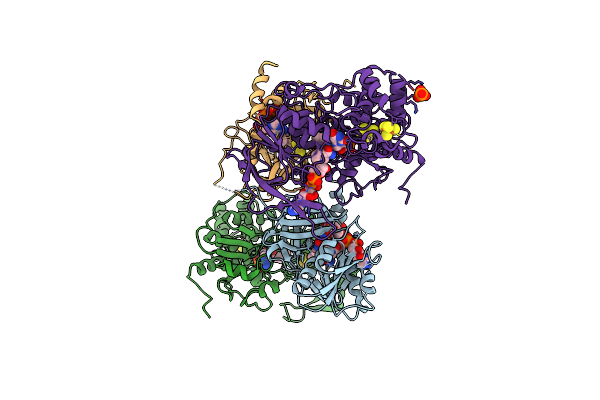
Insights Into Flavin-Based Electron Bifurcation Via The Nadh-Dependent Reduced Ferredoxin-Nadp Oxidoreductase Structure
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-07-08 Classification: OXIDOREDUCTASE Ligands: FES, FAD, SF4, PO4 |
 |
Insights Into Flavin-Based Electron Bifurcation Via The Nadh-Dependent Reduced Ferredoxin-Nadp Oxidoreductase Structure
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-07-08 Classification: OXIDOREDUCTASE Ligands: FES, FAD, NAD, SF4, PO4 |
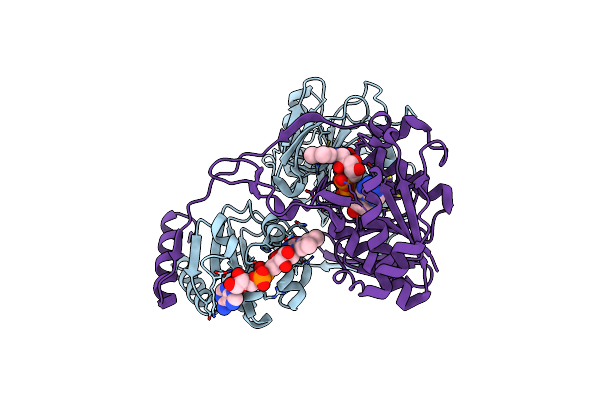 |
Electron Transferring Flavoprotein Of Acidaminococcus Fermentans: Towards A Mechanism Of Flavin-Based Electron Bifurcation
Organism: Acidaminococcus fermentans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-01-15 Classification: ELECTRON TRANSPORT Ligands: FAD, CL |
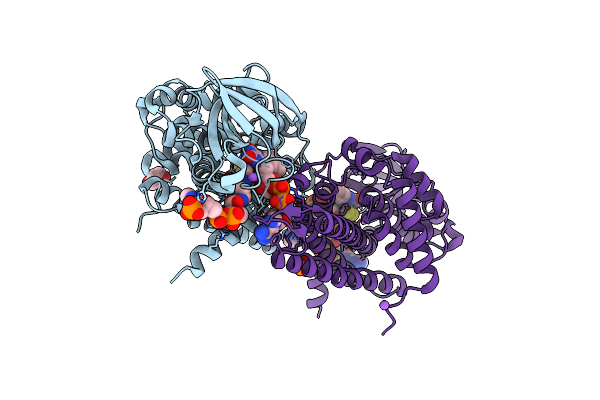 |
Electron Transferring Flavoprotein Of Acidaminococcus Fermentans: Towards A Mechanism Of Flavin-Based Electron Bifurcation
Organism: Acidaminococcus fermentans
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2014-01-15 Classification: ELECTRON TRANSPORT Ligands: FAD, COS, NA, PDO, PO4 |
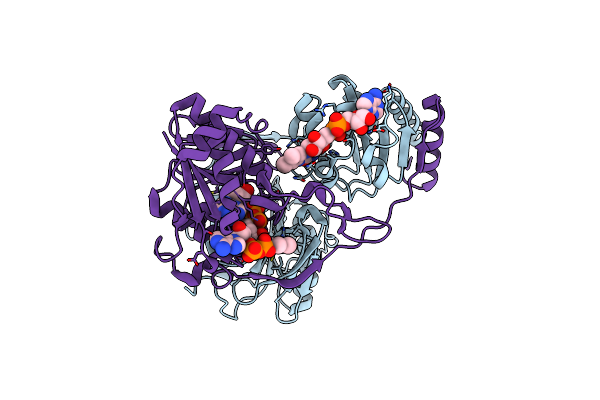 |
Electron Transferring Flavoprotein Of Acidaminococcus Fermentans: Towards A Mechanism Of Flavin-Based Electron Bifurcation
Organism: Acidaminococcus fermentans
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-01-15 Classification: ELECTRON TRANSPORT Ligands: FAD, CL, NAD |
 |
Structure Of The Rhodococcus Haloalkane Dehalogenase Mutant With Enhanced Enantioselectivity
Organism: Rhodococcus rhodochrous
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2008-09-16 Classification: HYDROLASE |
 |
Organism: Trichosurus vulpecula
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-11-20 Classification: TRANSPORT PROTEIN |
 |
Organism: Trichosurus vulpecula
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-11-20 Classification: TRANSPORT PROTEIN Ligands: ZN, CL, IPA |
 |
Crystal Structure Of The Possum Milk Whey Lipocalin Trichosurin At Ph 4.6 With Bound 4-Ethylphenol
Organism: Trichosurus vulpecula
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2007-11-20 Classification: TRANSPORT PROTEIN Ligands: ZN, CL, ETY, IPA |

