Search Count: 9
 |
Crystal Structure Of Sclam, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA, GOL |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With 1,3-Beta-D-Cellotriosyl-Glucose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With 1,3-Beta-D-Cellobiosyl-Cellobiose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Cellotriose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose And A Cellobiose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Cellohexaose, Presenting A 1,3-Beta-D-Cellobiosyl-Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Sclam E144S Mutant, A Non-Specific Endo-Beta-1,3(4)-Glucanase From Family Gh16, Co-Crystallized With Laminarihexaose, Presenting A Laminaribiose And A Glucose At Active Site
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-02-10 Classification: HYDROLASE Ligands: PO4, GLC, CA |
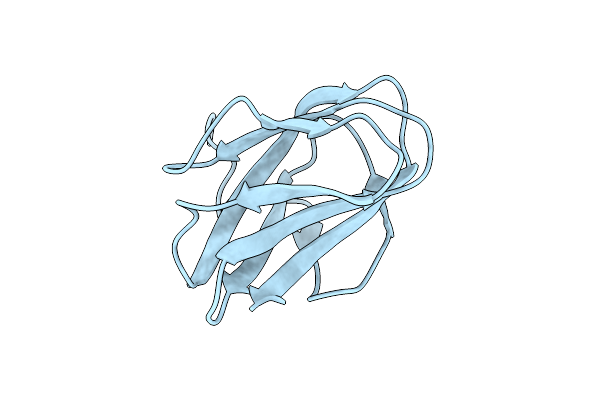 |
Structure Of Cbm_E1, A Novel Carbohydrate-Binding Module Found By Sugar Cane Soil Metagenome
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-09-21 Classification: SUGAR BINDING PROTEIN |
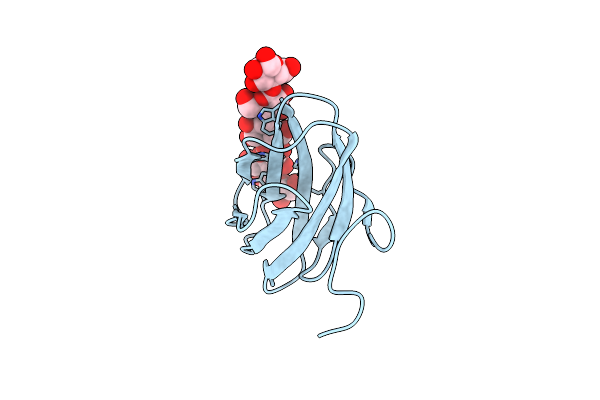 |
Structure Of Cbm_E1, A Novel Carbohydrate-Binding Module Found By Sugar Cane Soil Metagenome, Complexed With Cellopentaose
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-09-21 Classification: SUGAR BINDING PROTEIN |
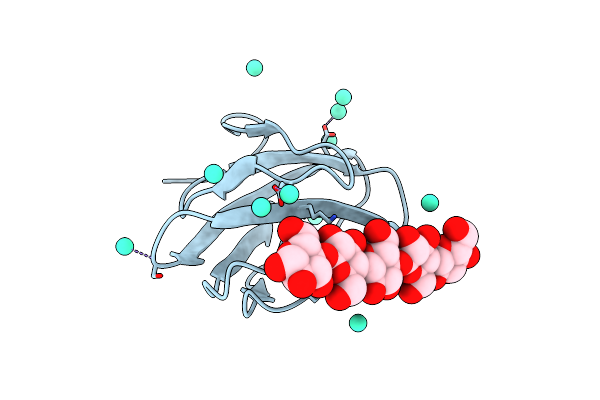 |
Structure Of Cbm_E1, A Novel Carbohydrate-Binding Module Found By Sugar Cane Soil Metagenome, Complexed With Cellopentaose And Gadolinium Ion
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-09-21 Classification: SUGAR BINDING PROTEIN Ligands: GD |

