Search Count: 132
 |
Organism: Rhodococcus rhodochrous
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Organism: Rhodococcus jostii rha1
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-10-02 Classification: OXIDOREDUCTASE Ligands: HEM, ANN, GOL |
 |
Organism: Rhodococcus jostii rha1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-10-02 Classification: OXIDOREDUCTASE Ligands: HEM, EGM, GOL |
 |
Organism: Rhodococcus jostii rha1
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-10-02 Classification: OXIDOREDUCTASE Ligands: HEM, TWO, GOL |
 |
Organism: Staphylococcaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-05-01 Classification: HYDROLASE |
 |
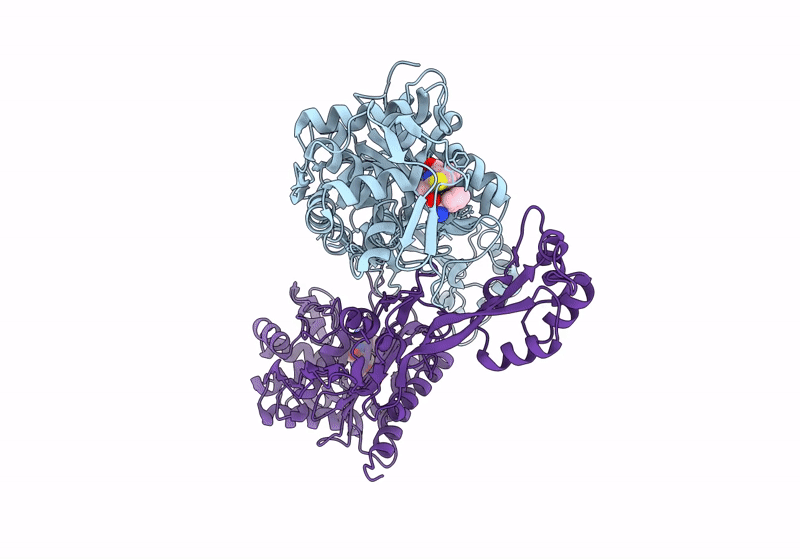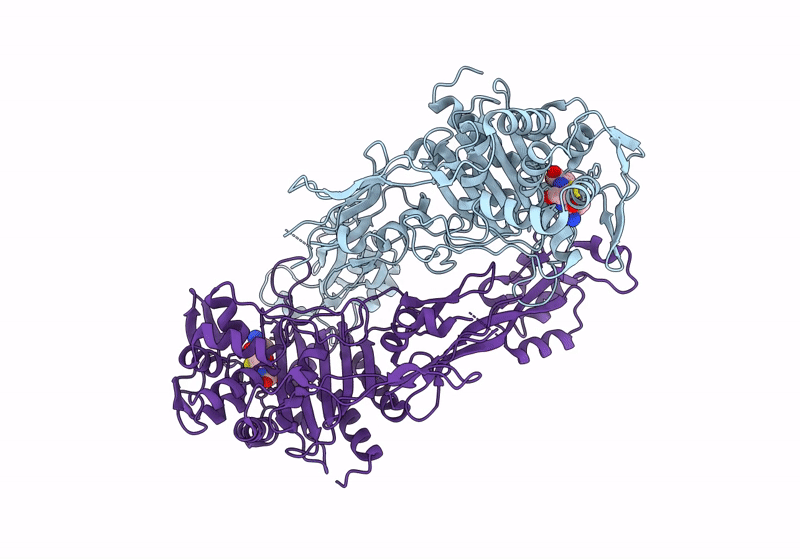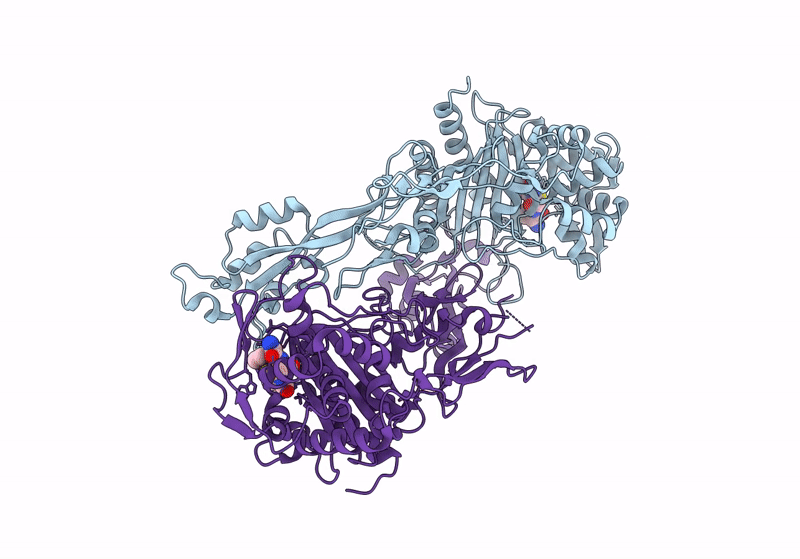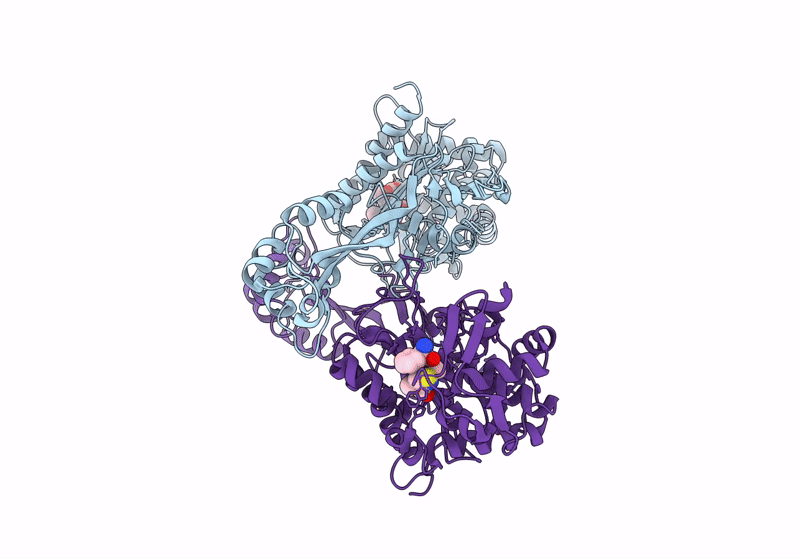
Structure Of The Monofunctional Staphylococcus Aureus Pbp1 In Its Beta-Lactam (Oxacillin) Inhibited Form
Organism: Staphylococcaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-05-01 Classification: HYDROLASE/INHIBITOR,ANTIBIOTIC Ligands: 1S6 |
 |
Structure Of The Monofunctional Staphylococcus Aureus Pbp1 In Its Beta-Lactam (Cephalexin) Inhibited Form
Organism: Staphylococcaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-05-01 Classification: HYDROLASE/INHIBITOR,ANTIBIOTIC Ligands: 63U |
 |
Structure Of The Monofunctional Staphylococcus Aureus Pbp1 In Its Beta-Lactam (Ertapenem) Inhibited Form
Organism: Staphylococcaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-05-01 Classification: HYDROLASE/INHIBITOR,ANTIBIOTIC Ligands: 1RG |
 |
Organism: Rhodococcus jostii (strain rha1)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-04-06 Classification: OXIDOREDUCTASE Ligands: FE, CA, 8RU |
 |
Organism: Cupriavidus metallidurans ch34
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-12-15 Classification: OXIDOREDUCTASE Ligands: EDO, GLU, GLY, FES, FE2, GOL |
 |
Organism: Comamonas testosteroni (strain dsm 14576 / kf-1)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2021-12-15 Classification: OXIDOREDUCTASE Ligands: FE2, FES, TLA, SRT, EDO |
 |
Organism: Comamonas testosteroni (strain dsm 14576 / kf-1)
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2021-12-15 Classification: OXIDOREDUCTASE Ligands: FE2, FES, PHT |
 |
Organism: Comamonas testosteroni (strain dsm 14576 / kf-1)
Method: X-RAY DIFFRACTION Resolution:3.07 Å Release Date: 2021-12-15 Classification: OXIDOREDUCTASE Ligands: FE2, FES, UB7 |
 |
Organism: Pseudoalteromonas luteoviolacea
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-10-06 Classification: BIOSYNTHETIC PROTEIN Ligands: BTB, ACT, EDO |
 |
Organism: Pseudoalteromonas luteoviolacea
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-10-06 Classification: BIOSYNTHETIC PROTEIN Ligands: BTB, ACT, EJ1, GOL, EDO, AE3 |
 |
Organism: Pseudoalteromonas luteoviolacea
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-10-06 Classification: BIOSYNTHETIC PROTEIN Ligands: BTB, EDO, 4VI |
 |
Organism: Sphingobium sp. (strain nbrc 103272 / syk-6)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-05-12 Classification: OXIDOREDUCTASE Ligands: CO |
 |
Organism: Sphingobium sp. (strain nbrc 103272 / syk-6)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-05-12 Classification: OXIDOREDUCTASE Ligands: V5M, DMS, CO, MG |
 |
Crystal Structure Of Lignostilbene Bound To Co-Lsd4 From Sphingobium Sp. Strain Syk-6
Organism: Sphingobium sp. (strain nbrc 103272 / syk-6)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-05-12 Classification: OXIDOREDUCTASE Ligands: CO, V5P, DMS, MG |
 |
Crystal Structure Of Vanillin Bound To Co-Lsd4 From Sphingobium Sp. Strain Syk-6
Organism: Sphingobium sp. (strain nbrc 103272 / syk-6)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-05-12 Classification: OXIDOREDUCTASE Ligands: V55, DMS, ACT, CO, MG |

