Search Count: 8
 |
N-Acetylglucosamine 6-Phosphate Dehydratase: Inhibited 6-Phosphogluconic Acid State Of Nags
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4, 6PG |
 |
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
N-Acetylglucosamine 6-Phosphate Dehydratase: Glcnac6P Substrate-Bound State Of Nags
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2025-02-19 Classification: SUGAR BINDING PROTEIN Ligands: SO4, 16G |
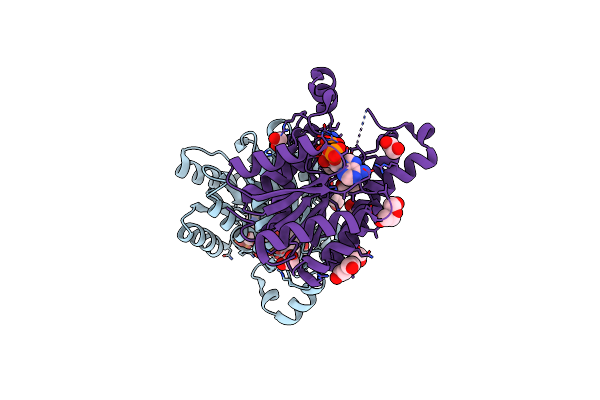 |
Promiscuous Reductase Lugoii Catalyzes Keto-Reduction At C1 During Lugdunomycin Biosynthesis
Organism: Streptomyces sp. ql37
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2020-09-16 Classification: ANTIBIOTIC Ligands: NDP, EDO, PEG |
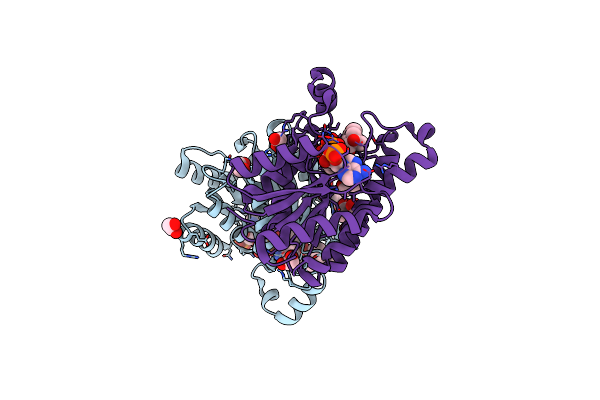 |
Promiscuous Reductase Lugoii Catalyzes Keto-Reduction At C1 During Lugdunomycin Biosynthesis
Organism: Streptomyces sp. ql37
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2020-09-16 Classification: ANTIBIOTIC Ligands: EDO, NAP, P7K |
 |
Promiscuous Reductase Lugoii Catalyzes Keto-Reduction At C1 During Lugdunomycin Biosynthesis
Organism: Streptomyces sp. ql37
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2020-09-16 Classification: ANTIBIOTIC Ligands: NAP, P7Q, EDO |
 |
Promiscuous Reductase Lugoii Catalyzes Keto-Reduction At C1 During Lugdunomycin Biosynthesis
Organism: Streptomyces sp. ql37
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2020-09-16 Classification: ANTIBIOTIC Ligands: EDO, PEG, SO4 |
 |
Organism: Streptomyces leeuwenhoekii
Method: SOLUTION NMR Release Date: 2015-10-07 Classification: CELL INVASION |

