Search Count: 80
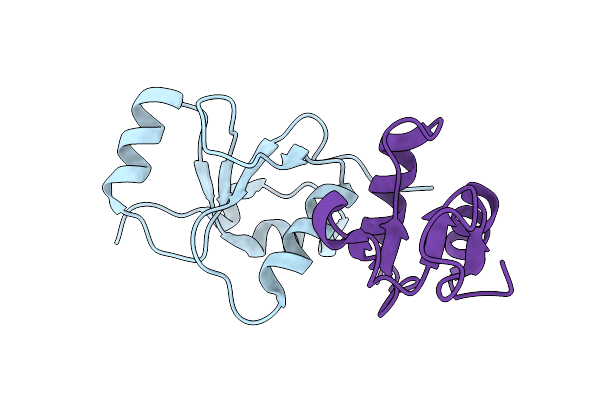 |
Structure Of The Complex Of Human Dna Ligase Iii-Alpha And Xrcc1 Brct Domains
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-12-02 Classification: DNA BINDING PROTEIN/LIGASE |
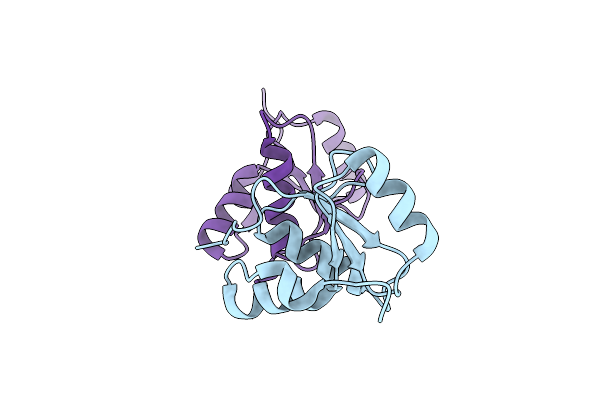 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2020-12-02 Classification: DNA BINDING PROTEIN |
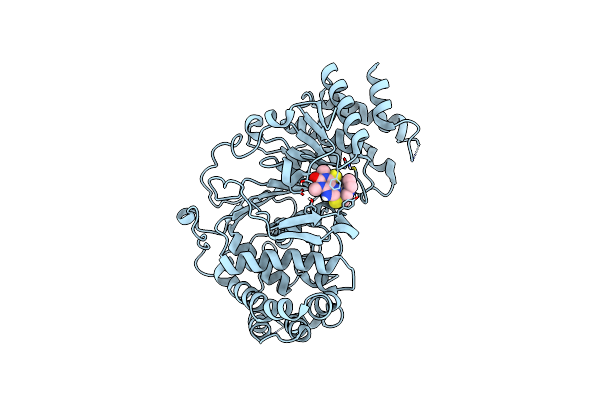 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-03-18 Classification: hydrolase/hydrolase inhibitor Ligands: M0M |
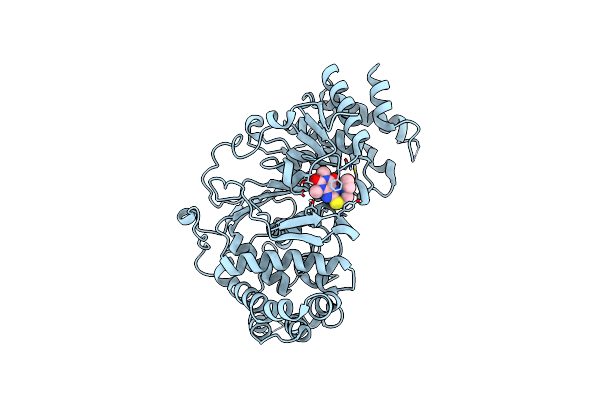 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-03-18 Classification: hydrolase/hydrolase inhibitor Ligands: M0P |
 |
Structure Of T7 Dna Polymerase Bound To A Primer/Template Dna And A Peptide That Mimics The C-Terminal Tail Of The Primase-Helicase
Organism: Enterobacteria phage t7, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-03-04 Classification: TRANSFERASE/DNA BINDING PROTEIN/DNA Ligands: TTP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-12-25 Classification: hydrolase/hydrolase inhibitor Ligands: M0S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-12-25 Classification: hydrolase/hydrolase inhibitor Ligands: M0V |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-12-25 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: M1D |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-12-25 Classification: hydrolase/hydrolase inhibitor Ligands: M0P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-12-25 Classification: hydrolase/hydrolase inhibitor Ligands: M0M |
 |
Organism: Enterobacteria phage t7, Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:4.80 Å Release Date: 2016-12-07 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2016-11-16 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Crystal Structure Of Monomeric Human Apoptosis-Inducing Factor With E413A/R422A/R430A Mutations
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-11-16 Classification: OXIDOREDUCTASE Ligands: FAD, EPE, GOL |
 |
Crystal Structure Of The Catalytic Domain Of Rat Poly (Adp-Ribose) Glycohydrolase
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-05-23 Classification: HYDROLASE |
 |
Crystal Structure Of The Catalytic Domain Of Rat Poly (Adp-Ribose) Glycohydrolase Bound To Adp-Hpd
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2012-05-23 Classification: HYDROLASE Ligands: A1R |
 |
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-03-16 Classification: HYDROLASE INHIBITOR Ligands: SO4 |
 |
Crystal Structure Of The Streptococcus Pyogenes Nad+ Glycohydrolase Spn In Complex With Ifs, The Immunity Factor For Spn
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2011-03-02 Classification: HYDROLASE/HYDROLASE INHIBITOR |
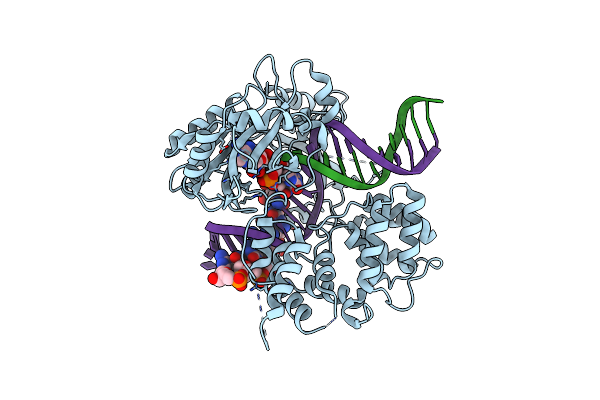 |
Human Dna Ligase Iii Recognizes Dna Ends By Dynamic Switching Between Two Dna Bound States
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-07-14 Classification: LIGASE/DNA Ligands: AMP |
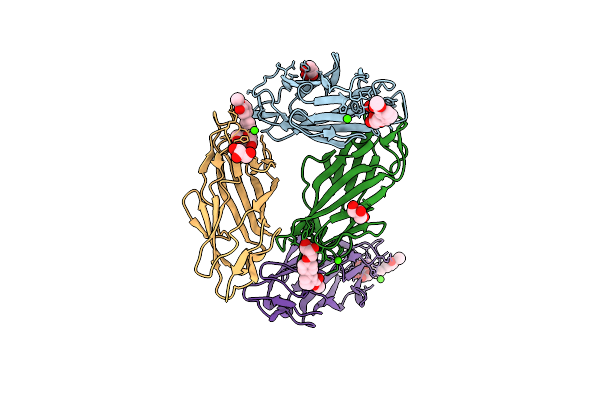 |
Crystal Structure Of Fimh Lectin Domain Bound To Biphenyl Mannoside Meta-Methyl Ester.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-06-16 Classification: carbohydrate binding protein Ligands: ZH1, GOL, CA, CL |
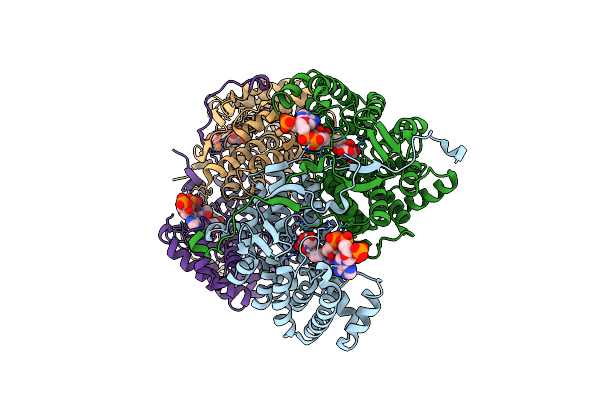 |
The Structure Of The Binary Complex Of Citryl Dethia Coa And Citrate Synthase From The Thermophilic Archaeonthermoplasma Acidophilum
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2008-09-30 Classification: TRANSFERASE Ligands: SDX |

