Search Count: 18
 |
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: ZN, ACT, EDO, SO4 |
 |
Thermodynamic, Crystallographic And Computational Studies Of Non Mammalian Fatty Acid Binding To Bovine B-Lactoglobulin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-06-20 Classification: TRANSPORT PROTEIN Ligands: EW8 |
 |
Thermodynamic, Crystallographic And Computational Studies Of Non Mammalian Fatty Acid Binding To Bovine B-Lactoglobulin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-06-20 Classification: TRANSPORT PROTEIN Ligands: X90 |
 |
Thermodynamic, Crystallographic And Computational Studies Of Non Mammalian Fatty Acid Binding To Bovine B-Lactoglobulin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-06-20 Classification: TRANSPORT PROTEIN Ligands: F15, CL |
 |
Thermodynamic, Crystallographic And Computational Studies Of Non Mammalian Fatty Acid Binding To Bovine B-Lactoglobulin
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-06-20 Classification: TRANSPORT PROTEIN Ligands: TDA |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ZN, ACT, EDO |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With N-Hydroxynaphthalene-1-Carboxamide
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: 8GK, ZN, EDO, NA, PGE |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With (E)-N-Hydroxy-3-(Naphthalen-1-Yl)Prop-2-Enamide
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ACT, ZN, EDO, 8SQ, PGE |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With (2S)-2,6-Diamino-N-Hydroxyhexanamide
Organism: Bacillus cereus atcc 14579, Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: 8SZ, ZN, CIT, EDO, PGE |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With (2S)-2-Amino-5-(Diaminomethylideneamino)-N-Hydroxypentanamide
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ACT, ZN, AHL, CIT |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With Acetazolamide
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ZN, ACT, AZM, PGE |
 |
Crystal Structure Of The Polysaccharide Deacetylase Bc1974 From Bacillus Cereus In Complex With Thiametg
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: ZN, ACT, NHT, PGE, EDO, CIT |
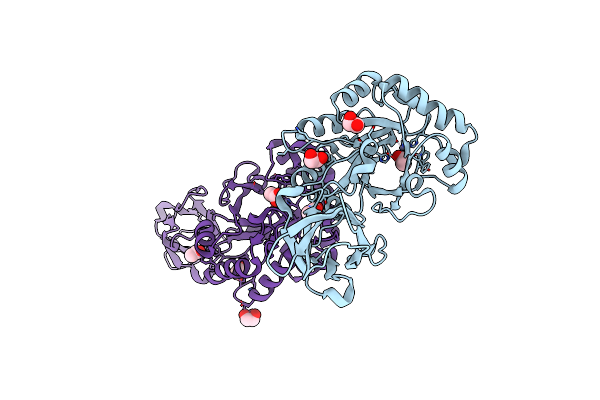 |
Crystal Structure Of The Putative Polysaccharide Deacetylase Ba0330 From Bacillus Anthracis
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2015-04-08 Classification: HYDROLASE Ligands: ACT, ZN, EDO |
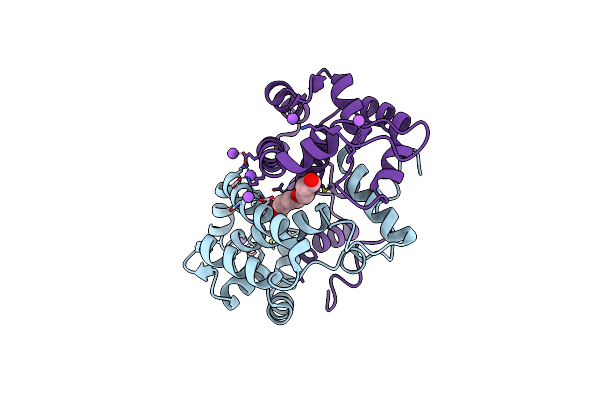 |
Crystal Structure Of Odorant Binding Protein 48 From Anopheles Gambiae (Agamobp48) With Peg
Organism: Anopheles gambiae
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-10-16 Classification: TRANSPORT PROTEIN Ligands: PEU, NA |
 |
Crystal Structure Of Odorant Binding Protein 48 From Anopheles Gambiae At 3.3 Angstrom Resolution
Organism: Anopheles gambiae
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2013-10-16 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Odorant Binding Protein 1 From Anopheles Gambiae (Agamobp1) With Deet (N,N-Diethyl-Meta-Toluamide) And Peg
Organism: Anopheles gambiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-06-08 Classification: TRANSPORT PROTEIN Ligands: MG, MOH, DE3, PEU, CL |
 |
Organism: Proteus hauseri
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2010-04-14 Classification: HYDROLASE Ligands: SO4, MPD, TRS |
 |
Crystal Structure Of Fab198, An Efficient Protector Of Acetylcholine Receptor Against Myasthenogenic Antibodies
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2001-09-26 Classification: IMMUNE SYSTEM |

